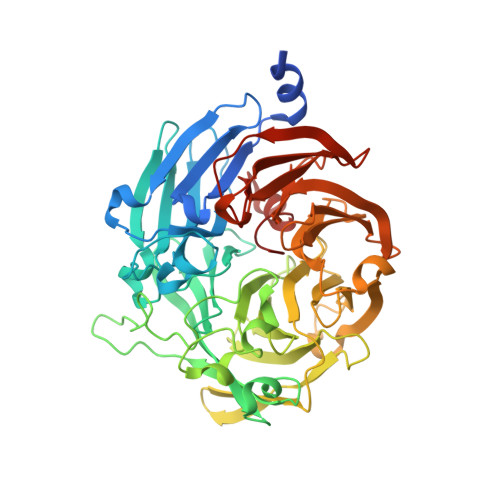
The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum (pmTcDH), activated by crystals soaking with 1 mM CuCl2
Varfolomeeva, L.A., Solovieva, A.Y., Shipkov, N.S., Dergousova, N.I., Boyko, K.M., Tikhonova, T.V., Popov, V.O.To be published.