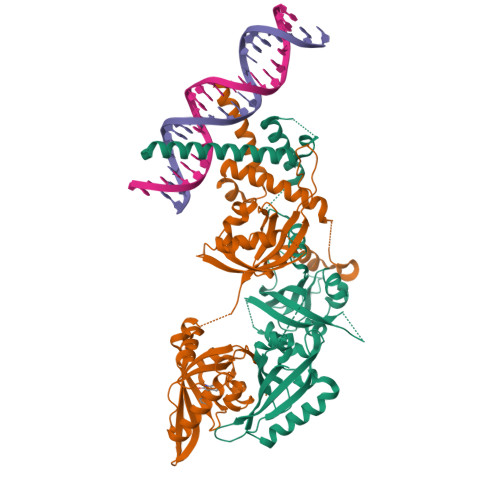
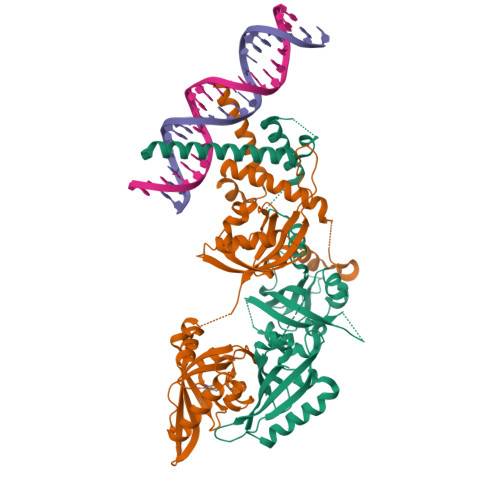
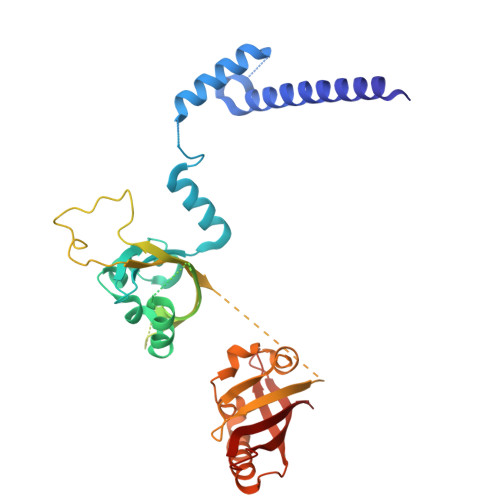
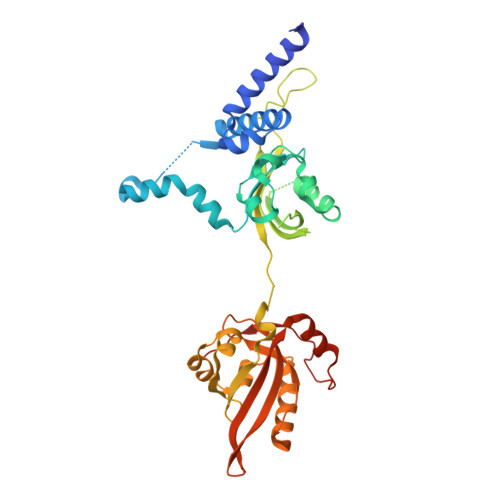
Structural basis for the ligand-dependent activation of heterodimeric AHR-ARNT complex.
Diao, X., Shang, Q., Guo, M., Huang, Y., Zhang, M., Chen, X., Liang, Y., Sun, X., Zhou, F., Zhuang, J., Liu, S.J., Vogel, C.F.A., Rastinejad, F., Wu, D.(2025) Nat Commun 16: 1282-1282
- PubMed: 39900897
- DOI: https://doi.org/10.1038/s41467-025-56574-7
- Primary Citation of Related Structures:
8XS6, 8XS7, 8XS8, 8XS9, 8XSA, 8XSB - PubMed Abstract:
The aryl hydrocarbon receptor (AHR) possesses an extraordinary capacity to sense and respond to a wide range of small-molecule ligands, ranging from polycyclic aromatic hydrocarbons to endogenous compounds. Upon ligand binding, AHR translocates from the cytoplasm to nucleus, forming a transcriptionally active complex with aryl hydrocarbon receptor nuclear translocator (ARNT), for DNA binding and initiation of gene expression programs that include cellular detoxification pathways and immune responses. Here, we examine the molecular mechanisms governing AHR's high-affinity binding and activation by a diverse group of ligands. Crystal structures of the AHR-ARNT-DNA complexes, bound with each of six established AHR ligands, including Tapinarof, 6-formylindolo[3,2-b]carbazole (FICZ), benzo[a]pyrene (BaP), β-naphthoflavone (BNF), Indigo and Indirubin, reveal an unconventional mode of subunit assembly with intimate association between the PAS-B domains of AHR and ARNT. AHR's PAS-B domain utilizes eight conserved residues whose dynamic rearrangements account for the ability to bind to ligands through hydrophobic and π-π interactions. Our findings further reveal the structural underpinnings of a ligand-driven activation mechanism, whereby a segment of the AHR protein undergoes a structural transition from chaperone engagement to ARNT heterodimer stabilization, to generate the transcriptionally competent assembly. Our results provide key information for the future development of AHR-targeting drugs.
Organizational Affiliation:
State Key Laboratory of Microbial Technology, Shandong University, Qingdao, China.