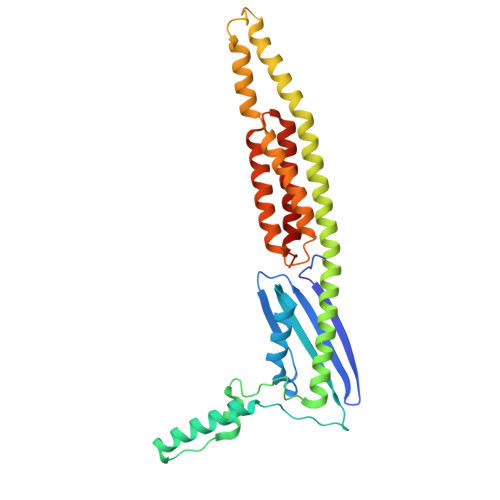
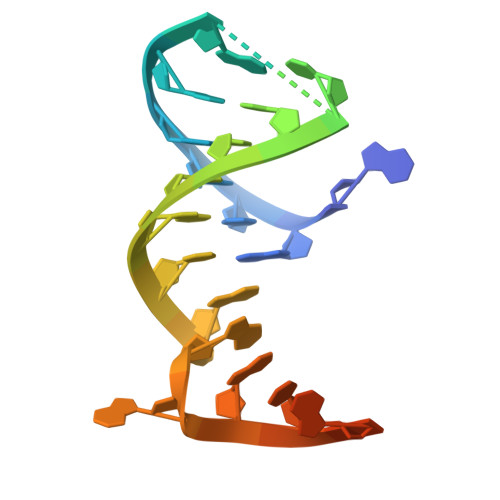
Structural insights into RNA cleavage by a novel family of bacterial RNases.
Wu, R., Ingle, S., Barnes, S.A., Dahlin, H.R., Khamrui, S., Xiang, Y., Shi, Y., Bechhofer, D.H., Lazarus, M.B.(2024) Nucleic Acids Res
- PubMed: 39180400
- DOI: https://doi.org/10.1093/nar/gkae717
- Primary Citation of Related Structures:
8VER, 8VES - PubMed Abstract:
Processing of RNA is a key regulatory mechanism for all living systems. Escherichia coli protein YicC belongs to the well-conserved YicC family and has been identified as a novel ribonuclease. Here, we report a 2.8-Å-resolution crystal structure of the E. coli YicC apo protein and a 3.2-Å-cryo-EM structure of YicC bound to an RNA substrate. The apo YicC forms a dimer of trimers with a large open channel. In the RNA-bound form, the top trimer of YicC rotates nearly 70° and closes the RNA substrate inside the cavity to form a clamshell-pearl conformation that resembles no other known RNases. The structural information combined with mass spectrometry and biochemical data identified cleavage on the upstream side of an RNA hairpin. Mutagenesis studies demonstrated that the previously uncharacterized domain, DUF1732, is critical in both RNA binding and catalysis. These studies shed light on the mechanism of the previously unexplored YicC RNase family.
Organizational Affiliation:
Department of Pharmacological Sciences, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.