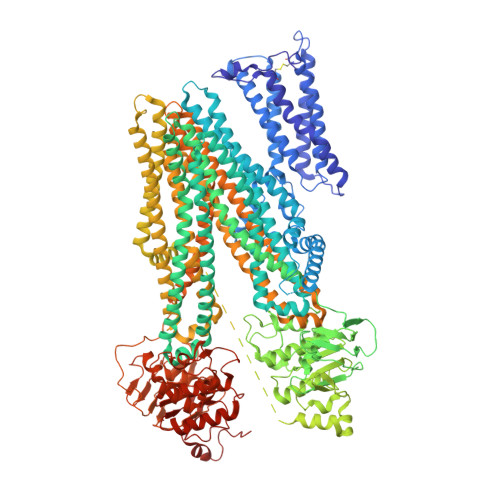
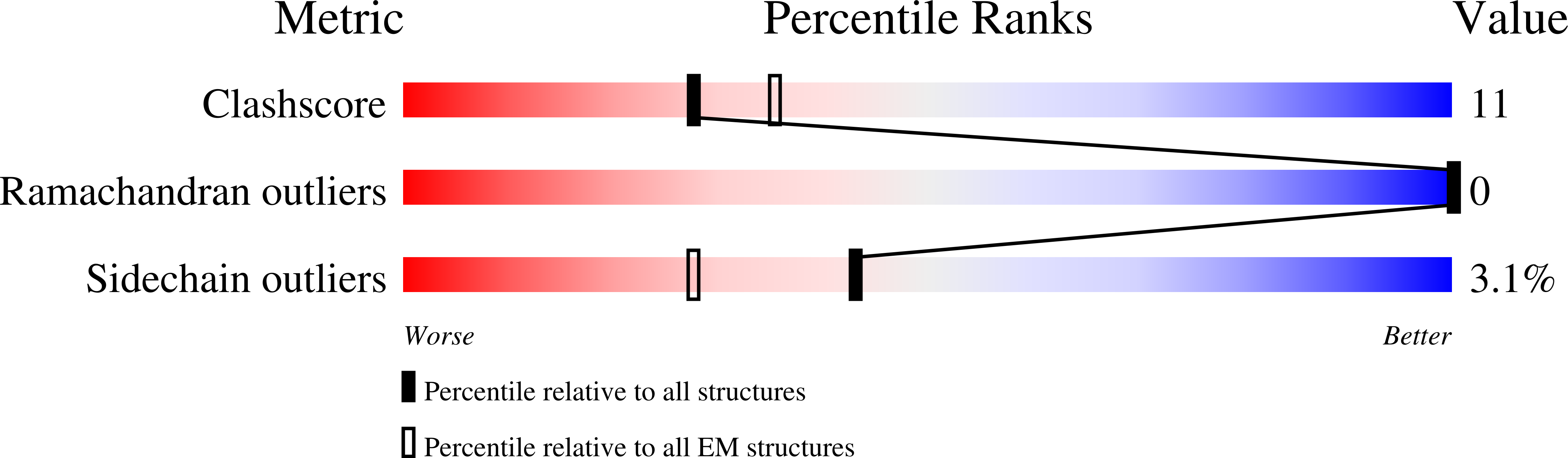Structural basis for the modulation of MRP2 activity by phosphorylation and drugs.
Mazza, T., Roumeliotis, T.I., Garitta, E., Drew, D., Rashid, S.T., Indiveri, C., Choudhary, J.S., Linton, K.J., Beis, K.(2024) Nat Commun 15: 1983-1983
- PubMed: 38438394
- DOI: https://doi.org/10.1038/s41467-024-46392-8
- Primary Citation of Related Structures:
8RQ3, 8RQ4 - PubMed Abstract:
Multidrug resistance-associated protein 2 (MRP2/ABCC2) is a polyspecific efflux transporter of organic anions expressed in hepatocyte canalicular membranes. MRP2 dysfunction, in Dubin-Johnson syndrome or by off-target inhibition, for example by the uricosuric drug probenecid, elevates circulating bilirubin glucuronide and is a cause of jaundice. Here, we determine the cryo-EM structure of rat Mrp2 (rMrp2) in an autoinhibited state and in complex with probenecid. The autoinhibited state exhibits an unusual conformation for this class of transporter in which the regulatory domain is folded within the transmembrane domain cavity. In vitro phosphorylation, mass spectrometry and transport assays show that phosphorylation of the regulatory domain relieves this autoinhibition and enhances rMrp2 transport activity. The in vitro data is confirmed in human hepatocyte-like cells, in which inhibition of endogenous kinases also reduces human MRP2 transport activity. The drug-bound state reveals two probenecid binding sites that suggest a dynamic interplay with autoinhibition. Mapping of the Dubin-Johnson mutations onto the rodent structure indicates that many may interfere with the transition between conformational states.
Organizational Affiliation:
Department of Life Sciences, Imperial College London, SW7 2AZ, London, UK.