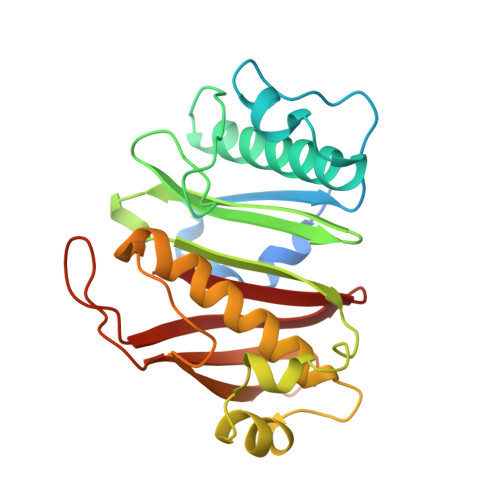
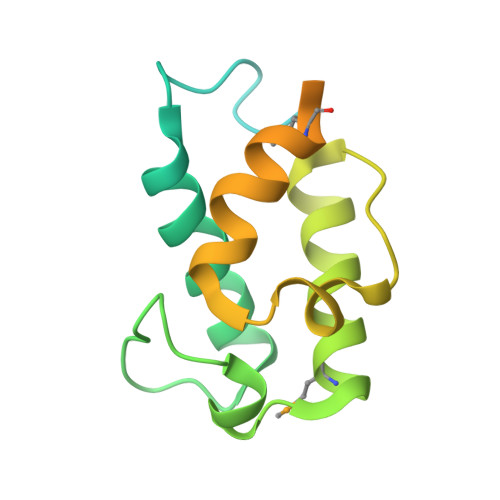
Catalytic Cycle of Type II 4'-Phosphopantetheinyl Transferases
Gavalda, S., Faille, A., Fioccola, S., Nguyen, M.C., Carivenc, C., Rottier, K., Rufin, Y., Saitta, S., Czaplicki, G., Guilhot, C., Chalut, C., Brut, M., Mourey, L., Pedelacq, J.D.(2024) ACS Catal 14: 8561-8575