A novel pathological mutant reveals the role of torsional flexibility in the serpin breach in adoption of an aggregation-prone intermediate.
Kamuda, K., Ronzoni, R., Majumdar, A., Guan, F.H.X., Irving, J.A., Lomas, D.A.(2024) FEBS J 291: 2937-2954
- PubMed: 38523412
- DOI: https://doi.org/10.1111/febs.17121
- Primary Citation of Related Structures:
8P4J, 8P4U - PubMed Abstract:
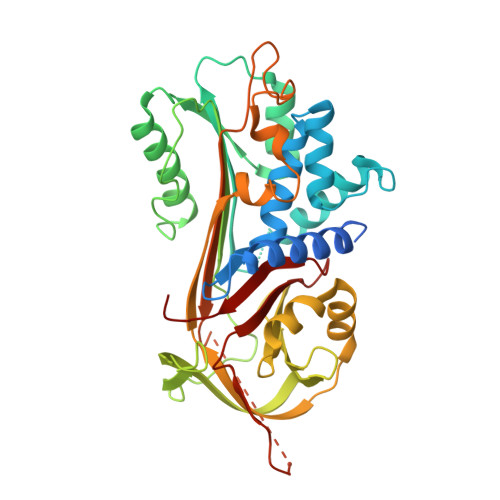
Mutants of alpha-1-antitrypsin cause the protein to self-associate and form ordered aggregates ('polymers') that are retained within hepatocytes, resulting in a predisposition to the development of liver disease. The associated reduction in secretion, and for some mutants, impairment of function, leads to a failure to protect lung tissue against proteases released during the inflammatory response and an increased risk of emphysema. We report here a novel deficiency mutation (Gly192Cys), that we name the Sydney variant, identified in a patient in heterozygosity with the Z allele (Glu342Lys). Cellular analysis revealed that the novel variant was mostly retained as insoluble polymers within the endoplasmic reticulum. The basis for this behaviour was investigated using biophysical and structural techniques. The variant showed a 40% reduction in inhibitory activity and a reduced stability as assessed by thermal unfolding experiments. Polymerisation involves adoption of an aggregation-prone intermediate and paradoxically the energy barrier for transition to this state was increased by 16% for the Gly192Cys variant with respect to the wild-type protein. However, with activation to the intermediate state, polymerisation occurred at a 3.8-fold faster rate overall. X-ray crystallography provided two crystal structures of the Gly192Cys variant, revealing perturbation within the 'breach' region with Cys192 in two different orientations: in one structure it faces towards the hydrophobic core while in the second it is solvent-exposed. This orientational heterogeneity was confirmed by PEGylation. These data show the critical role of the torsional freedom imparted by Gly192 in inhibitory activity and stability against polymerisation.
Organizational Affiliation:
Division of Medicine, UCL Respiratory, Rayne Institute, University College London, UK.