Structure of the Histidine Kinase CheA ATP-Binding domain in complex with compound QUI-SV-383
Adhav, A., Marina, A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chemotaxis protein CheA | A [auth B], B [auth A] | 189 | Thermotoga maritima | Mutation(s): 0 Gene Names: cheA, TM_0702 EC: 2.7.13.3 | 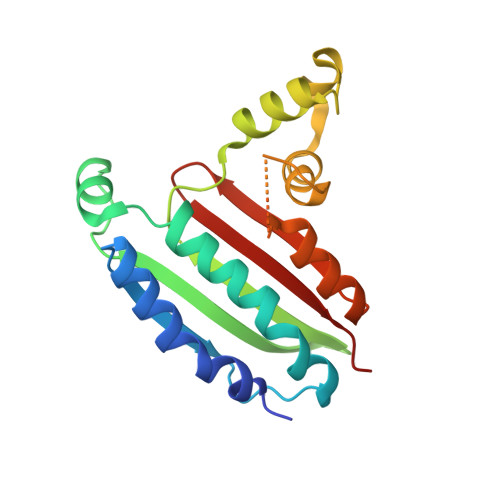 |
UniProt | |||||
Find proteins for Q56310 (Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8)) Explore Q56310 Go to UniProtKB: Q56310 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q56310 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| W6E (Subject of Investigation/LOI) Query on W6E | C [auth B], D [auth A] | 7-(1~{H}-1,2,3-triazol-5-yl)quinazolin-2-amine C10 H8 N6 BRODMNADNZNYMX-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 41.113 | α = 90 |
| b = 59.773 | β = 97.154 |
| c = 66.609 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| REFMAC | refinement |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| H2020 Marie Curie Actions of the European Commission | European Union | 765147 |