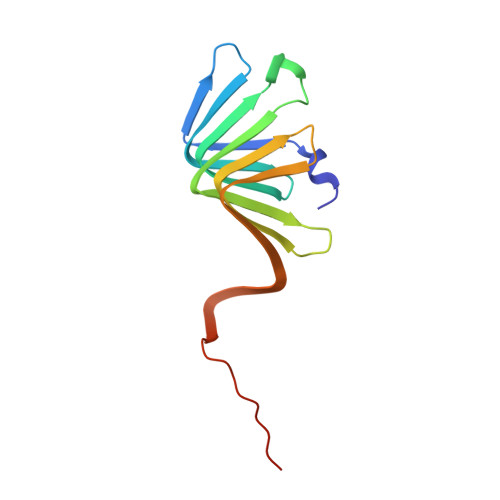
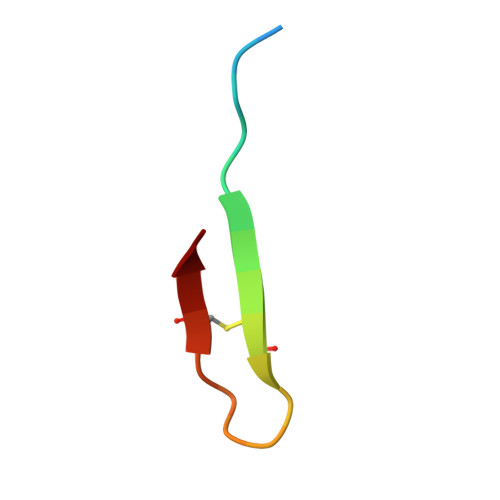
Discovery, characterization, and redesign of potent antimicrobial thanatin orthologs from Chinavia ubica and Murgantia histrionica targeting E. coli LptA.
Huynh, K., Kibrom, A., Donald, B.R., Zhou, P.(2023) J Struct Biol X 8: 100091-100091
- PubMed: 37416832
- DOI: https://doi.org/10.1016/j.yjsbx.2023.100091
- Primary Citation of Related Structures:
8GAJ, 8GAK, 8GAL - PubMed Abstract:
Podisus maculiventris thanatin has been reported as a potent antimicrobial peptide with antibacterial and antifungal activity. Its antibiotic activity has been most thoroughly characterized against E. coli and shown to interfere with multiple pathways, such as the lipopolysaccharide transport (LPT) pathway comprised of seven different Lpt proteins. Thanatin binds to E. coli LptA and LptD, thus disrupting the LPT complex formation and inhibiting cell wall synthesis and microbial growth. Here, we performed a genomic database search to uncover novel thanatin orthologs, characterized their binding to E. coli LptA using bio-layer interferometry, and assessed their antimicrobial activity against E. coli . We found that thanatins from Chinavia ubica and Murgantia histrionica bound tighter (by 3.6- and 2.2-fold respectively) to LptA and exhibited more potent antibiotic activity (by 2.1- and 2.8-fold respectively) than the canonical thanatin from P. maculiventris . We crystallized and determined the LptA-bound complex structures of thanatins from C. ubica (1.90 Å resolution), M. histrionica (1.80 Å resolution), and P. maculiventris (2.43 Å resolution) to better understand their mechanism of action. Our structural analysis revealed that residues A10 and I21 in C. ubica and M. histrionica thanatin are important for improving the binding interface with LptA, thus overall improving the potency of thanatin against E. coli . We also designed a stapled variant of thanatin that removes the need for a disulfide bond but retains the ability to bind LptA and antibiotic activity. Our discovery presents a library of novel thanatin sequences to serve as starting scaffolds for designing more potent antimicrobial therapeutics.
Organizational Affiliation:
Department of Biochemistry, Duke University School of Medicine, Durham, NC, United States.