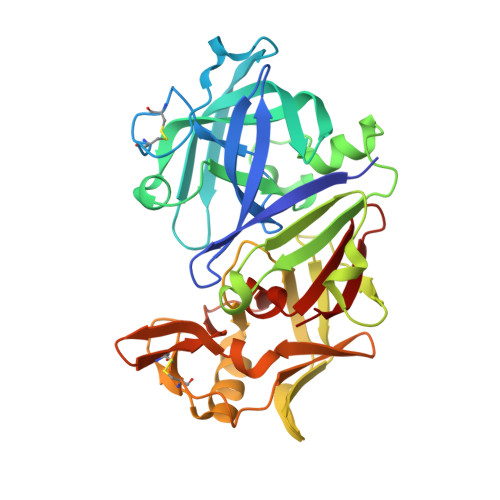
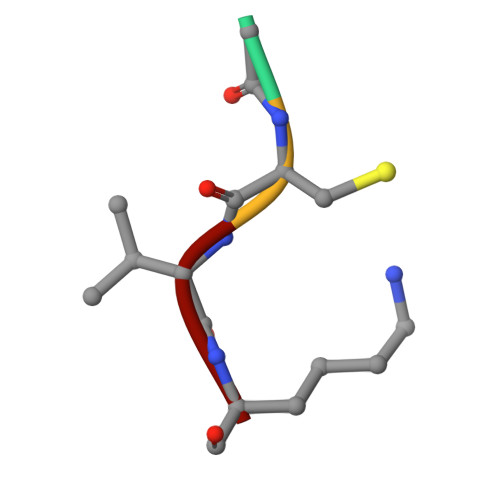
Aspartic protease inhibitors designed from computer-generated templates bind as predicted.
Ripka, A.S., Satyshur, K.A., Bohacek, R.S., Rich, D.H.(2001) Org Lett 3: 2309-2312
- PubMed: 11463303
- DOI: https://doi.org/10.1021/ol016090+
- Primary Citation of Related Structures:
8FXQ - PubMed Abstract:
[reaction: see text] Novel tripeptide-derived peptidomimetics 1, 7ab, and 8ab, inspired by templates generated by the structure-generating program GrowMol, were synthesized, shown to inhibit Rhizopus chinensis pepsin, and found by X-ray crystallography to bind to the enzyme in the GrowMol-predicted mode. Repetitive evaluation of the computer-generated templates for synthetic feasibility and optimal enzyme interactions led to the designed compounds.
Organizational Affiliation:
School of Pharmacy, University of Wisconsin-Madison, 777 Highland Avenue, Madison, Wisconsin 53705, USA.