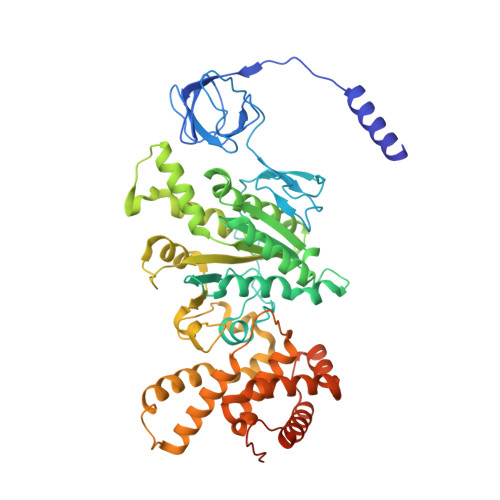
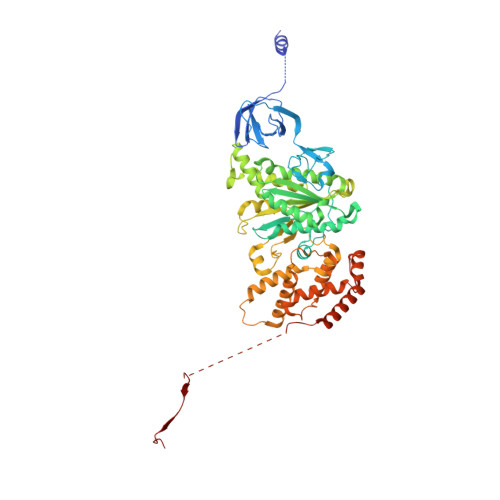
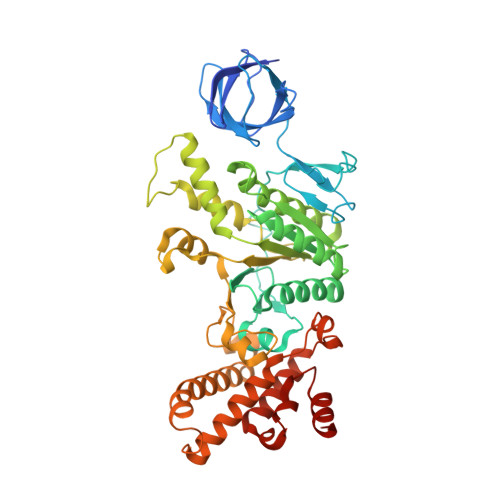
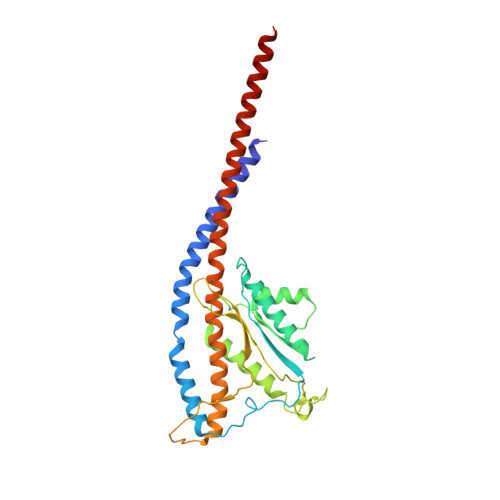
Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Wong, C.F., Saw, W.G., Basak, S., Sano, M., Ueno, H., Kerk, H.W., Litty, D., Ragunathan, P., Dick, T., Muller, V., Noji, H., Gruber, G.(2022) Antimicrob Agents Chemother 66: e0105622-e0105622
- PubMed: 36445139
- DOI: https://doi.org/10.1128/aac.01056-22
- Primary Citation of Related Structures:
7Y5A, 7Y5B, 7Y5C, 7Y5D - PubMed Abstract:
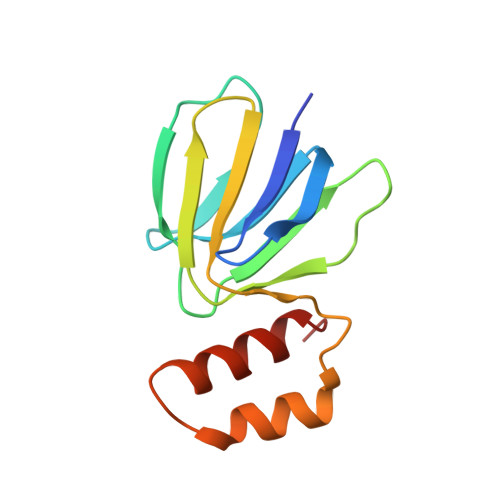
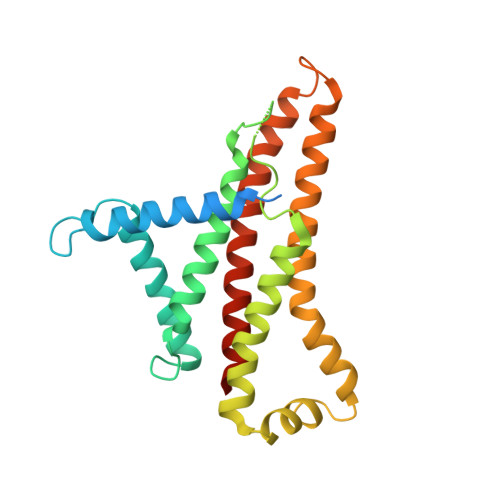
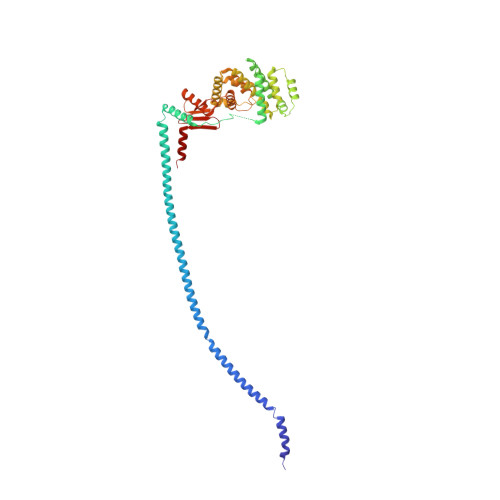
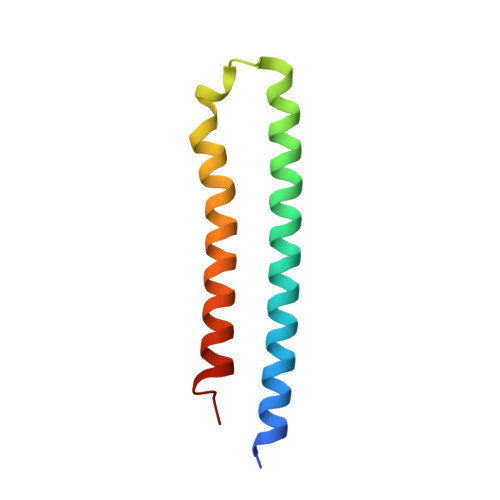
The F 1 F O -ATP synthase is required for the viability of tuberculosis (TB) and nontuberculous mycobacteria (NTM) and has been validated as a drug target. Here, we present the cryo-EM structures of the Mycobacterium smegmatis F 1 -ATPase and the F 1 F O -ATP synthase with different nucleotide occupation within the catalytic sites and visualize critical elements for latent ATP hydrolysis and efficient ATP synthesis. Mutational studies reveal that the extended C-terminal domain (αCTD) of subunit α is the main element for the self-inhibition mechanism of ATP hydrolysis for TB and NTM bacteria. Rotational studies indicate that the transition between the inhibition state by the αCTD and the active state is a rapid process. We demonstrate that the unique mycobacterial γ-loop and subunit δ are critical elements required for ATP formation. The data underline that these mycobacterium-specific elements of α, γ, and δ are attractive targets, providing a platform for the discovery of species-specific inhibitors.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological Universitygrid.59025.3b, Singapore, Singapore.