Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Luo, B., Zhang, C., Ling, X., Mukherjee, S., Jia, G., Xie, J., Jia, X., Liu, L., Baulin, E.F., Luo, Y., Jiang, L., Dong, H., Wei, X., Bujnicki, J.M., Su, Z.(2023) Nat Catal
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
This entry reflects an alternative modeling of the original data in: 7EZ0
(2023) Nat Catal
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| The Tet-S2 state with a pseudoknotted 4-way junction molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon (intron and 3'-exon) | A [auth N] | 434 | Tetrahymena thermophila | 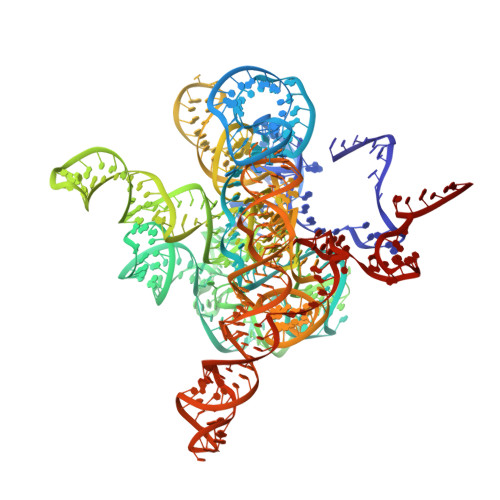 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| The Tet-S2 state with a pseudoknotted 4-way junction molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon (5'-exon) | B [auth A] | 25 | Tetrahymena thermophila | 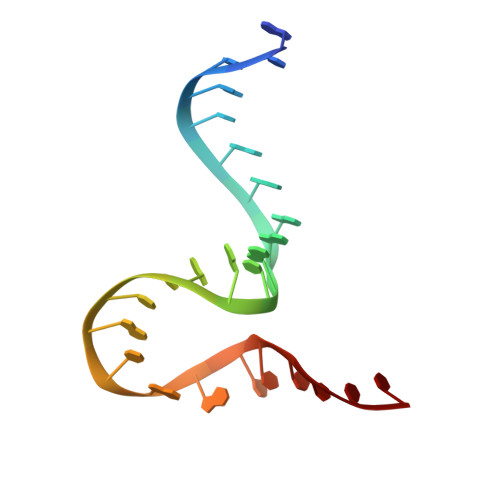 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SPD Query on SPD | EA [auth N] | SPERMIDINE C7 H19 N3 ATHGHQPFGPMSJY-UHFFFAOYSA-N |  | ||
| MG Query on MG | AA [auth N] BA [auth N] C [auth N] CA [auth N] D [auth N] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | RELION | 3.1 |
| MODEL REFINEMENT | Coot | 0.9.1 |
| MODEL REFINEMENT | PHENIX | 1.15 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Ministry of Science and Technology (MoST, China) | China | 2021YFA1301900 |
| Ministry of Science and Technology (MoST, China) | China | 2022YFC2303700 |
| National Natural Science Foundation of China (NSFC) | China | 32222040 |
| National Natural Science Foundation of China (NSFC) | China | 32070049 |
| Polish National Science Centre | Poland | 2017/26/A/NZ1/01083 |
| Polish National Science Centre | Poland | 2021/43/D/NZ1/03360 |
| European Molecular Biology Organization (EMBO) | European Union | -- |