A Novel Acyl-AcpM-Binding Protein Confers Intrinsic Sensitivity to Fatty Acid Synthase Type II Inhibitors in Mycobacterium smegmatis
Li, M., Huang, Q., Zhang, W., Cao, Y., Wang, Z., Zhao, Z., Zhang, X., Zhang, J.(2022) Front Microbiol 13: 846722-846722
- PubMed: 35444621
- DOI: https://doi.org/10.3389/fmicb.2022.846722
- Primary Citation of Related Structures:
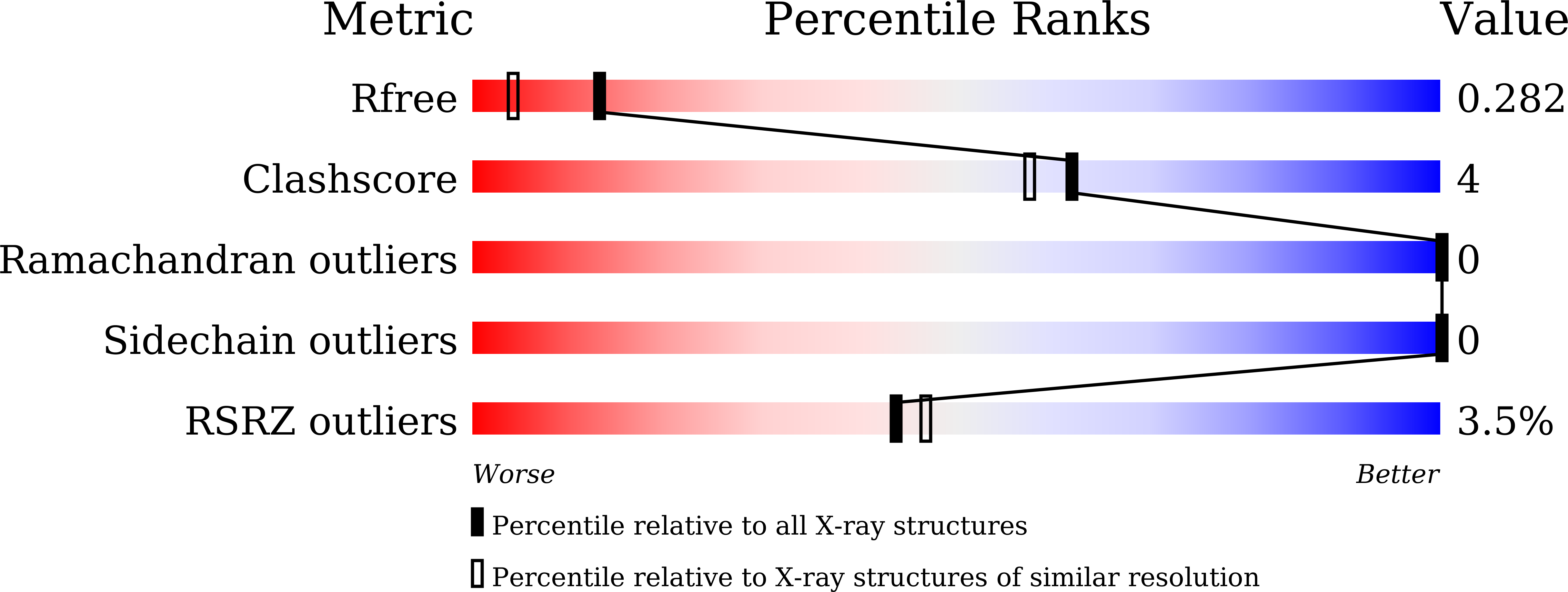
7WA9 - PubMed Abstract:
The fatty acid synthase type II (FAS-II) multienzyme system is the main target of drugs to inhibit mycolic acid synthesis in mycobacterium. Meromycolate extension acyl carrier protein (AcpM) serves as the carrier of fatty acyl chain shuttling among the individual FAS-II components during the progression of fatty acid elongation. In this paper, MSMEG_5634 in Mycobacterium smegmatis was determined to be a helix-grip structure protein with a deep hydrophobic pocket, preferring to form a complex with acyl-AcpM containing a fatty acyl chain at the C36-52 length, which is the medium product of FAS-II. MSMEG_5634 interacted with FAS-II components and presented relative accumulation at the cellular pole. By forming the MSMEG_5634/acyl-AcpM complex, which is free from FAS-II, MSMEG_5634 could transport acyl-AcpM away from FAS-II. Deletion of the MSMEG_5634 gene in M. smegmatis resulted in a mutant with decreased sensitivity to isoniazid and triclosan, two inhibitors of the FAS-II system. The isoniazid and triclosan sensitivity of this mutant could be restored by the ectopic expression of MSMEG_5634 or Rv0910, the MSMEG_5634 homologous protein in Mycobacterium tuberculosis H37Rv. These results suggest that MSMEG_5634 and its homologous proteins, forming a novel acyl-AcpM-binding protein family in mycobacterium, confer intrinsic sensitivity to FAS-II inhibitors.
Organizational Affiliation:
Key Laboratory of Cell Proliferation and Regulation Biology, Ministry of Education, Department of Biology, College of Life Sciences, Beijing Normal University, Beijing, China.