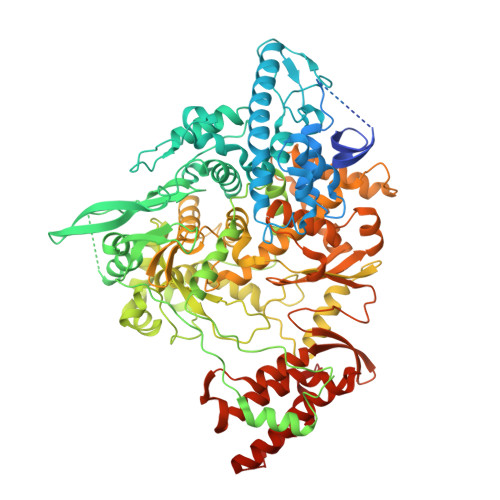
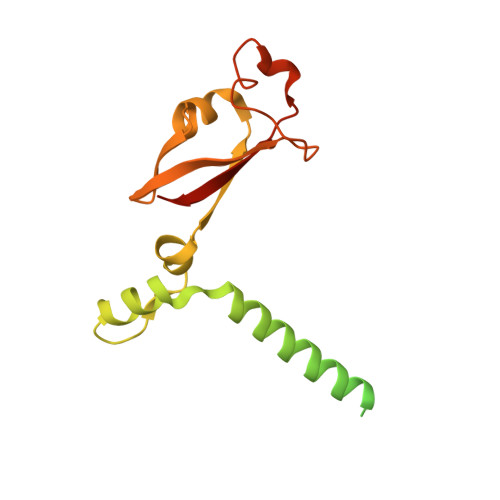
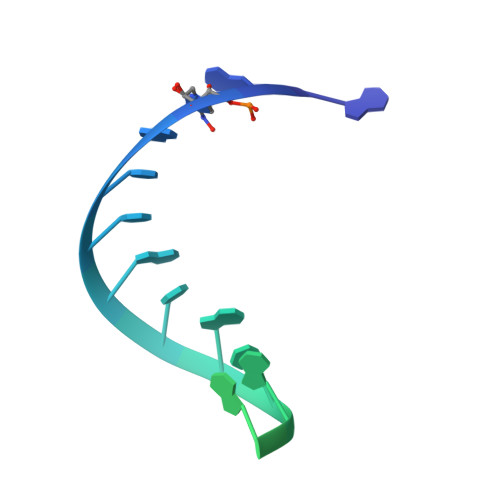
Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis.
Kabinger, F., Stiller, C., Schmitzova, J., Dienemann, C., Kokic, G., Hillen, H.S., Hobartner, C., Cramer, P.(2021) Nat Struct Mol Biol 28: 740-746
- PubMed: 34381216
- DOI: https://doi.org/10.1038/s41594-021-00651-0
- Primary Citation of Related Structures:
7OZU, 7OZV - PubMed Abstract:
Molnupiravir is an orally available antiviral drug candidate currently in phase III trials for the treatment of patients with COVID-19. Molnupiravir increases the frequency of viral RNA mutations and impairs SARS-CoV-2 replication in animal models and in humans. Here, we establish the molecular mechanisms underlying molnupiravir-induced RNA mutagenesis by the viral RNA-dependent RNA polymerase (RdRp). Biochemical assays show that the RdRp uses the active form of molnupiravir, β-D-N 4 -hydroxycytidine (NHC) triphosphate, as a substrate instead of cytidine triphosphate or uridine triphosphate. When the RdRp uses the resulting RNA as a template, NHC directs incorporation of either G or A, leading to mutated RNA products. Structural analysis of RdRp-RNA complexes that contain mutagenesis products shows that NHC can form stable base pairs with either G or A in the RdRp active center, explaining how the polymerase escapes proofreading and synthesizes mutated RNA. This two-step mutagenesis mechanism probably applies to various viral polymerases and can explain the broad-spectrum antiviral activity of molnupiravir.
Organizational Affiliation:
Max Planck Institute for Biophysical Chemistry, Department of Molecular Biology, Göttingen, Germany.