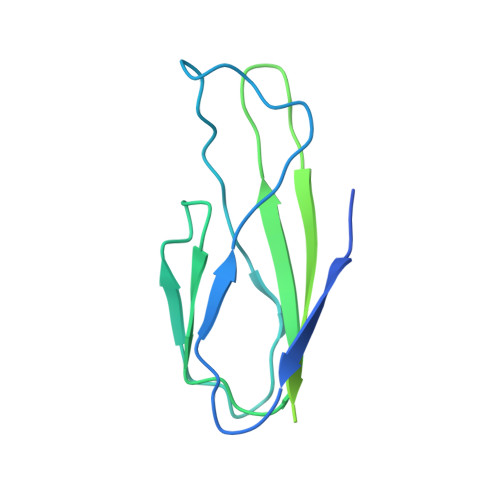
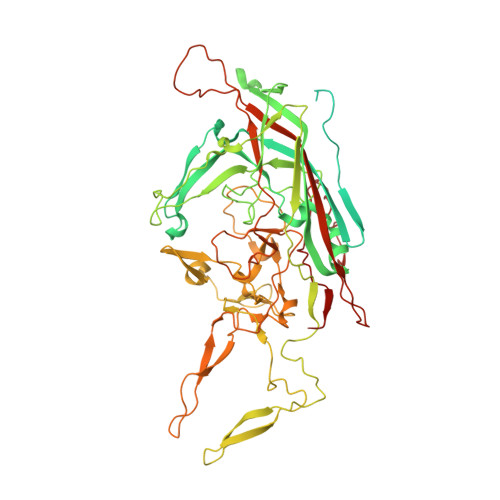
The Structure of an AAV5-AAVR Complex at 2.5 angstrom Resolution: Implications for Cellular Entry and Immune Neutralization of AAV Gene Therapy Vectors.
Silveria, M.A., Large, E.E., Zane, G.M., White, T.A., Chapman, M.S.(2020) Viruses 12
- PubMed: 33218165
- DOI: https://doi.org/10.3390/v12111326
- Primary Citation of Related Structures:
7KP3, 7KPN - PubMed Abstract:
Adeno-Associated Virus is the leading vector for gene therapy. Although it is the vector for all in vivo gene therapies approved for clinical use by the US Food and Drug Administration, its biology is still not yet fully understood. It has been shown that different serotypes of AAV bind to their cellular receptor, AAVR, in different ways. Previously we have reported a 2.4Å structure of AAV2 bound to AAVR that shows ordered structure for only one of the two AAVR domains with which AAV2 interacts. In this study we present a 2.5Å resolution structure of AAV5 bound to AAVR. AAV5 binds to the first polycystic kidney disease (PKD) domain of AAVR that was not ordered in the AAV2 structure. Interactions of AAV5 with AAVR are analyzed in detail, and the implications for AAV2 binding are explored through molecular modeling. Moreover, we find that binding sites for the antibodies ADK5a, ADK5b, and 3C5 on AAV5 overlap with the binding site of AAVR. These insights provide a structural foundation for development of gene therapy agents to better evade immune neutralization without disrupting cellular entry.
Organizational Affiliation:
Department of Biochemistry, University of Missouri, Columbia, MO 65211, USA.