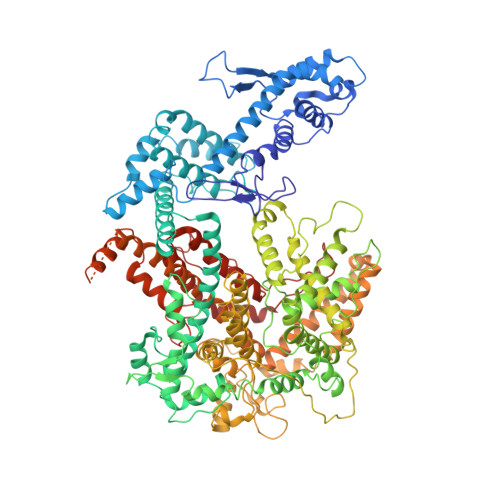
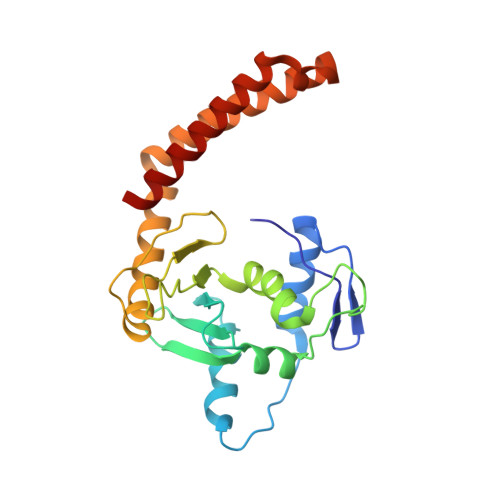
A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity.
Meeske, A.J., Jia, N., Cassel, A.K., Kozlova, A., Liao, J., Wiedmann, M., Patel, D.J., Marraffini, L.A.(2020) Science 369: 54-59
- PubMed: 32467331
- DOI: https://doi.org/10.1126/science.abb6151
- Primary Citation of Related Structures:
6VRB, 6VRC - PubMed Abstract:
The CRISPR RNA (crRNA)-guided nuclease Cas13 recognizes complementary viral transcripts to trigger the degradation of both host and viral RNA during the type VI CRISPR-Cas antiviral response. However, how viruses can counteract this immunity is not known. We describe a listeriaphage (ϕLS46) encoding an anti-CRISPR protein (AcrVIA1) that inactivates the type VI-A CRISPR system of Listeria seeligeri Using genetics, biochemistry, and structural biology, we found that AcrVIA1 interacts with the guide-exposed face of Cas13a, preventing access to the target RNA and the conformational changes required for nuclease activation. Unlike inhibitors of DNA-cleaving Cas nucleases, which cause limited immunosuppression and require multiple infections to bypass CRISPR defenses, a single dose of AcrVIA1 delivered by an individual virion completely dismantles type VI-A CRISPR-mediated immunity.
Organizational Affiliation:
Laboratory of Bacteriology, The Rockefeller University, New York, NY 10065, USA.