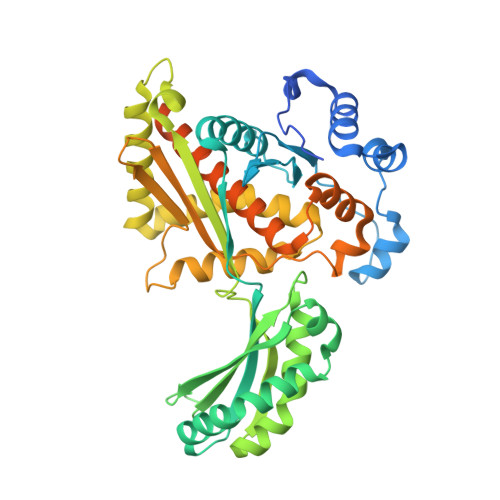
Structural and Functional Characterization of Dynamic Oligomerization in Burkholderia cenocepacia HMG-CoA Reductase.
Peacock, R.B., Hicks, C.W., Walker, A.M., Dewing, S.M., Lewis, K.M., Abboud, J.C., Stewart, S.W.A., Kang, C., Watson, J.M.(2019) Biochemistry 58: 3960-3970
- PubMed: 31469273
- DOI: https://doi.org/10.1021/acs.biochem.9b00494
- Primary Citation of Related Structures:
6P7K - PubMed Abstract:
The enzyme 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase (HMGR), in most organisms, catalyzes the four-electron reduction of the thioester ( S )-HMG-CoA to the primary alcohol ( R )-mevalonate, utilizing NADPH as the hydride donor. In some organisms, including the opportunistic lung pathogen Burkholderia cenocepacia , it catalyzes the reverse reaction, utilizing NAD + as a hydride acceptor in the oxidation of mevalonate. B. cenocepacia HMGR has been previously shown to exist as an ensemble of multiple non-additive oligomeric states, each with different levels of enzymatic activity, suggesting that the enzyme exhibits characteristics of the morpheein model of allostery. We have characterized a number of factors, including pH, substrate concentration, and enzyme concentration, that modulate the structural transitions that influence the interconversion among the multiple oligomers. We have also determined the crystal structure of B. cenocepacia HMGR in the hexameric state bound to coenzyme A and ADP. This hexameric assembly provides important clues about how the transition among oligomers might occur, and why B. cenocepacia HMGR, unique among characterized HMGRs, exhibits morpheein-like behavior.
Organizational Affiliation:
Department of Chemistry and Biochemistry , Gonzaga University , Spokane , Washington 99258 , United States.