Characterization of a family I inorganic pyrophosphatase from Legionella pneumophila Philadelphia 1.
Moorefield, J., Konuk, Y., Norman, J.O., Abendroth, J., Edwards, T.E., Lorimer, D.D., Mayclin, S.J., Staker, B.L., Craig, J.K., Barett, K.F., Barrett, L.K., Van Voorhis, W.C., Myler, P.J., McLaughlin, K.J.(2023) Acta Crystallogr F Struct Biol Commun 79: 257-266
- PubMed: 37728609
- DOI: https://doi.org/10.1107/S2053230X23008002
- Primary Citation of Related Structures:
6N1C - PubMed Abstract:
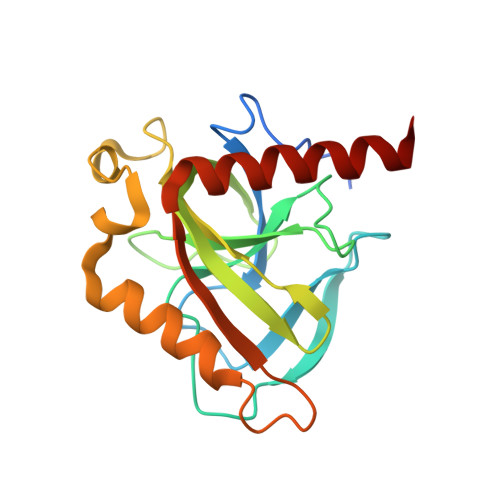
Inorganic pyrophosphate (PP i ) is generated as an intermediate or byproduct of many fundamental metabolic pathways, including DNA/RNA synthesis. The intracellular concentration of PP i must be regulated as buildup can inhibit many critical cellular processes. Inorganic pyrophosphatases (PPases) hydrolyze PP i into two orthophosphates (P i ), preventing the toxic accumulation of the PP i byproduct in cells and making P i available for use in biosynthetic pathways. Here, the crystal structure of a family I inorganic pyrophosphatase from Legionella pneumophila is reported at 2.0 Å resolution. L. pneumophila PPase (LpPPase) adopts a homohexameric assembly and shares the oligonucleotide/oligosaccharide-binding (OB) β-barrel core fold common to many other bacterial family I PPases. LpPPase demonstrated hydrolytic activity against a general substrate, with Mg 2+ being the preferred metal cofactor for catalysis. Legionnaires' disease is a severe respiratory infection caused primarily by L. pneumophila, and thus increased characterization of the L. pneumophila proteome is of interest.
Organizational Affiliation:
Department of Chemistry, Vassar College, 124 Raymond Avenue, Poughkeepsie, NY 12604, USA.