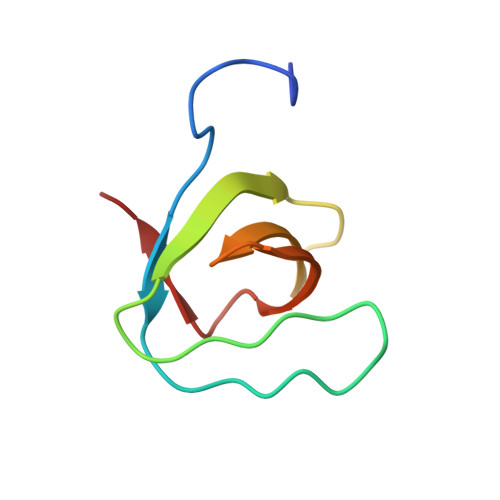
Lysozyme contamination facilitates crystallization of a heterotrimeric cortactin-Arg-lysozyme complex.
Liu, W., Macgrath, S.M., Koleske, A.J., Boggon, T.J.(2012) Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 154-158
- PubMed: 22297987
- DOI: https://doi.org/10.1107/S1744309111056132
- Primary Citation of Related Structures:
3ULR - PubMed Abstract:
Crystallization of contaminating proteins is a frequently encountered problem for macromolecular crystallographers. In this study, an attempt was made to obtain a binary cocrystal structure of the SH3 domain of cortactin and a 17-residue peptide from the Arg nonreceptor tyrosine kinase encompassing a PxxPxxPxxP (PxxP1) motif. However, cocrystals could only be obtained in the presence of trace amounts of a contaminating protein. A structure solution obtained by molecular replacement followed by ARP/wARP automatic model building allowed a 'sequence-by-crystallography' approach to discover that the contaminating protein was lysozyme. This 1.65 Å resolution crystal structure determination of a 1:1:1 heterotrimeric complex of Arg, cortactin and lysozyme thus provides an unusual `caveat emptor' warning of the dangers that underpurified proteins harbor for macromolecular crystallographers.
Organizational Affiliation:
Department of Pharmacology, Yale University School of Medicine, New Haven, CT 06520, USA.