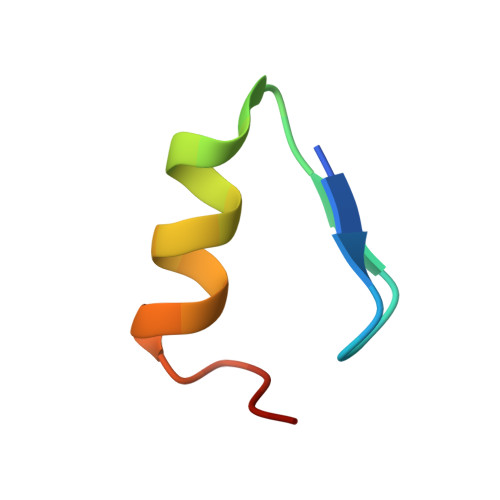
NMR structure verifies the eponymous zinc finger domain of transcription factor ZNF750.
Rua, A.J., Whitehead 3rd, R.D., Alexandrescu, A.T.(2023) J Struct Biol X 8: 100093-100093
- PubMed: 37655311
- DOI: https://doi.org/10.1016/j.yjsbx.2023.100093
- Primary Citation of Related Structures:
8SXM - PubMed Abstract:
ZNF750 is a nuclear transcription factor that activates skin differentiation and has tumor suppressor roles in several cancers. Unusually, ZNF750 has only a single zinc-finger (ZNF) domain, Z*, with an amino acid sequence that differs markedly from the CCHH family consensus. Because of its sequence differences Z* is classified as degenerate, presumed to have lost the ability to bind the zinc ion required for folding. AlphaFold predicts an irregular structure for Z* with low confidence. Low confidence predictions are often inferred to be intrinsically disordered regions of proteins, which would be the case if Z* did not bind Zn 2+ . We use NMR and CD spectroscopy to show that a 25-51 segment of ZNF750 corresponding to the Z* domain folds into a well-defined antiparallel ββα tertiary structure with a pM dissociation constant for Zn 2+ and a thermal stability >80 °C. Of three alternative Zn 2+ ligand sets, Z* uses a CCHC rather than the expected CCHH ligating motif. The switch in the last ligand maintains the folding topology and hydrophobic core of the classical ZNF motif. CCHC ZNFs are typically associated with protein-protein interactions, raising the possibility that ZNF750 interacts with DNA through other proteins rather than directly. The structure of Z* provides context for understanding the function of the domain and its cancer-associated mutations. We expect other ZNFs currently classified as degenerate could be CCHC-type structures like Z*.
Organizational Affiliation:
Department of Molecular and Cellular Biology, University of Connecticut, United States.