Discovery, Characterization, and Bioactivity of the Achromonodins: Lasso Peptides Encoded by Achromobacter .
Carson, D.V., Zhang, Y., So, L., Cheung-Lee, W.L., Cartagena, A.J., Darst, S.A., Link, A.J.(2023) J Nat Prod 86: 2448-2456
- PubMed: 37870195
- DOI: https://doi.org/10.1021/acs.jnatprod.3c00536
- Primary Citation of Related Structures:
8SVB - PubMed Abstract:
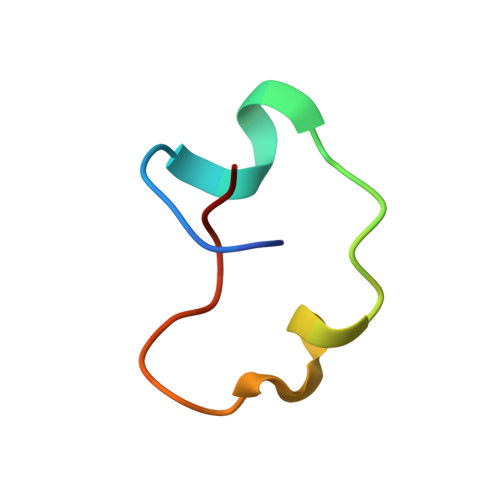
Through genome mining efforts, two lasso peptide biosynthetic gene clusters (BGCs) within two different species of Achromobacter , a genus that contains pathogenic organisms that can infect patients with cystic fibrosis, were discovered. Using gene-refactored BGCs in E. coli , these lasso peptides, which were named achromonodin-1 and achromonodin-2, were heterologously expressed. Achromonodin-1 is naturally encoded by certain isolates from the sputum of patients with cystic fibrosis. The NMR structure of achromonodin-1 was determined, demonstrating that it is a threaded lasso peptide with a large loop and short tail structure, reminiscent of previously characterized lasso peptides that inhibit RNA polymerase (RNAP). Achromonodin-1 inhibits RNAP in vitro and has potent, focused activity toward Achromobacter pulmonis , another isolate from the sputum of a cystic fibrosis patient. These efforts expand the repertoire of antimicrobial lasso peptides and provide insights into how Achromobacter isolates from certain ecological niches interact with each other.
Organizational Affiliation:
Department of Chemical and Biological Engineering, Princeton University, Princeton, New Jersey 08544, United States.