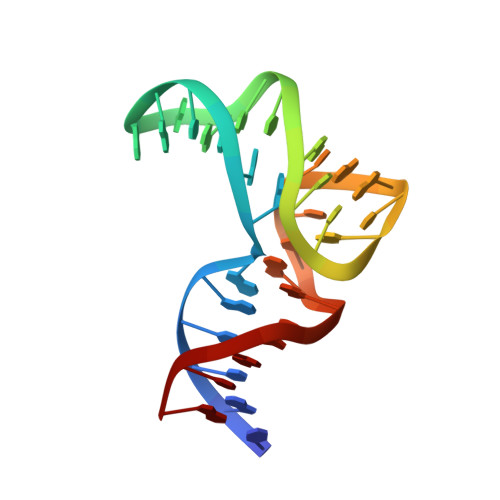
The 8-17 DNAzyme can operate in a single active structure regardless of metal ion cofactor.
Wieruszewska, J., Pawlowicz, A., Polomska, E., Pasternak, K., Gdaniec, Z., Andralojc, W.(2024) Nat Commun 15: 4218-4218
- PubMed: 38760331
- DOI: https://doi.org/10.1038/s41467-024-48638-x
- Primary Citation of Related Structures:
8OR8 - PubMed Abstract:
DNAzymes - synthetic enzymes made of DNA - have long attracted attention as RNA-targeting therapeutic agents. Yet, as of now, no DNAzyme-based drug has been approved, partially due to our lacking understanding of their molecular mode of action. In this work we report the solution structure of 8-17 DNAzyme bound to a Zn 2+ ion solved through NMR spectroscopy. Surprisingly, it turned out to be very similar to the previously solved Pb 2+ -bound form (catalytic domain RMSD = 1.28 Å), despite a long-standing literature consensus that Pb 2+ recruits a different DNAzyme fold than other metal ion cofactors. Our follow-up NMR investigations in the presence of other ions - Mg 2+ , Na + , and Pb 2+ - suggest that at DNAzyme concentrations used in NMR all these ions induce a similar tertiary fold. Based on these findings, we propose a model for 8-17 DNAzyme interactions with metal ions postulating the existence of only a single catalytically-active structure, yet populated to a different extent depending on the metal ion cofactor. Our results provide structural information on the 8-17 DNAzyme in presence of non-Pb 2+ cofactors, including the biologically relevant Mg 2+ ion.
Organizational Affiliation:
Institute of Bioorganic Chemistry, Polish Academy of Sciences, 61-704, Poznań, Noskowskiego, 12/14, Poland.