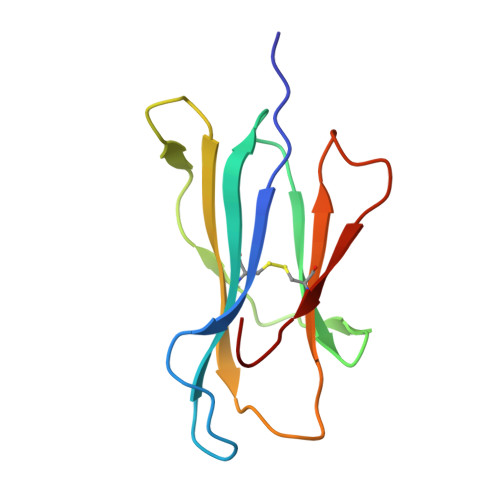
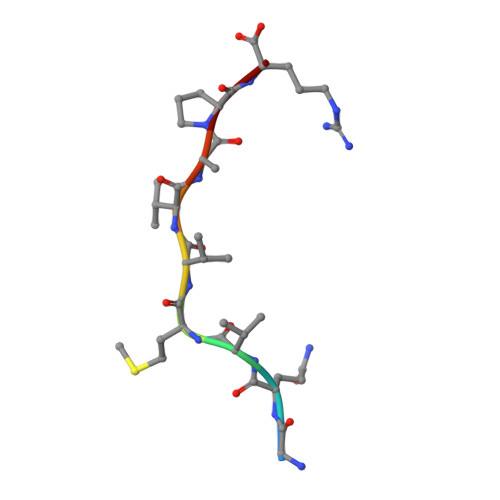
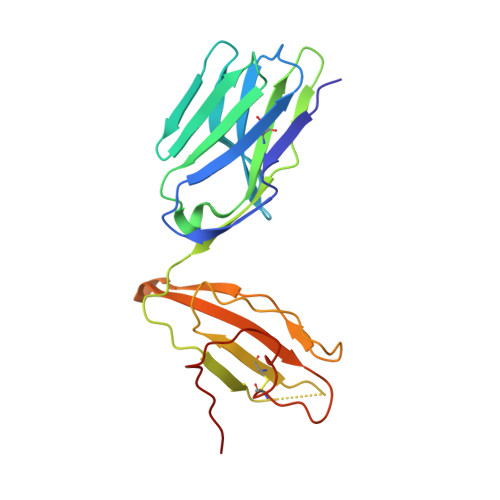
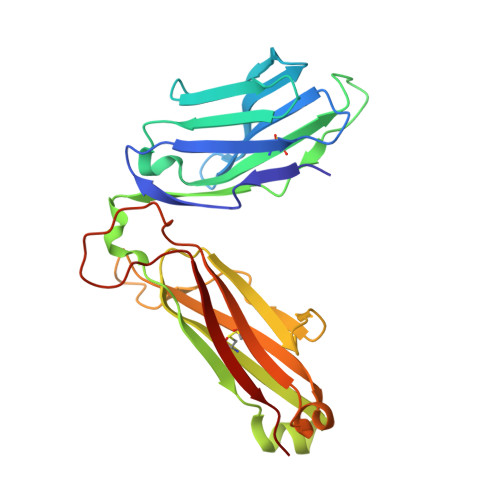
Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Yang, X., Garner, L.I., Zvyagin, I.V., Paley, M.A., Komech, E.A., Jude, K.M., Zhao, X., Fernandes, R.A., Hassman, L.M., Paley, G.L., Savvides, C.S., Brackenridge, S., Quastel, M.N., Chudakov, D.M., Bowness, P., Yokoyama, W.M., McMichael, A.J., Gillespie, G.M., Garcia, K.C.(2022) Nature 612: 771-777
- PubMed: 36477533
- DOI: https://doi.org/10.1038/s41586-022-05501-7
- Primary Citation of Related Structures:
7N2N, 7N2O, 7N2P, 7N2Q, 7N2R, 7N2S, 8CX4 - PubMed Abstract:
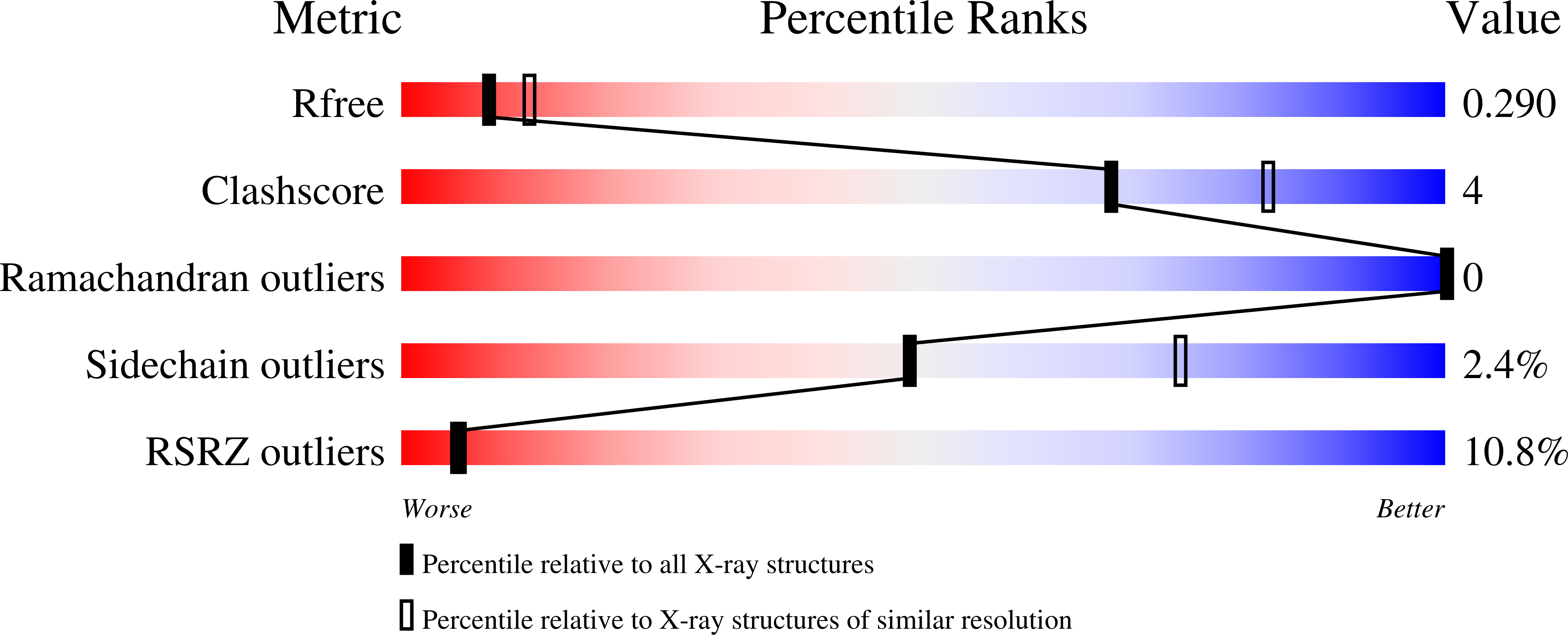
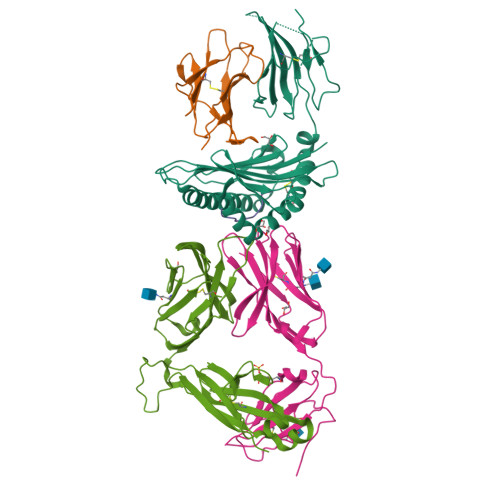
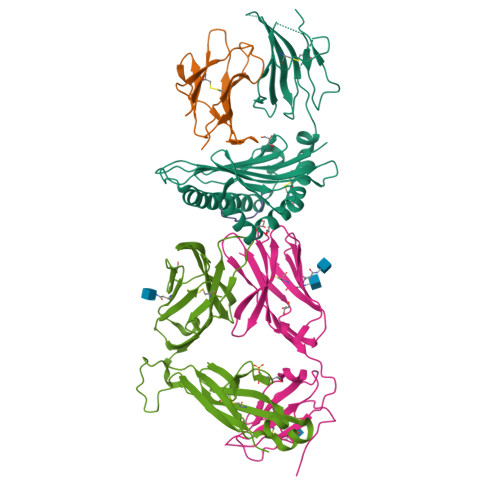
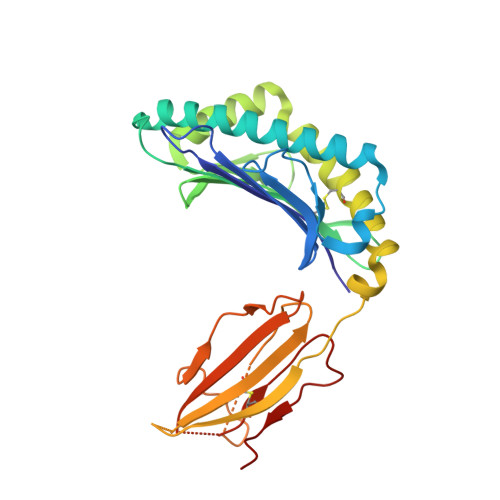
Human leucocyte antigen B*27 (HLA-B*27) is strongly associated with inflammatory diseases of the spine and pelvis (for example, ankylosing spondylitis (AS)) and the eye (that is, acute anterior uveitis (AAU)) 1 . How HLA-B*27 facilitates disease remains unknown, but one possible mechanism could involve presentation of pathogenic peptides to CD8 + T cells. Here we isolated orphan T cell receptors (TCRs) expressing a disease-associated public β-chain variable region-complementary-determining region 3β (BV9-CDR3β) motif 2-4 from blood and synovial fluid T cells from individuals with AS and from the eye in individuals with AAU. These TCRs showed consistent α-chain variable region (AV21) chain pairing and were clonally expanded in the joint and eye. We used HLA-B*27:05 yeast display peptide libraries to identify shared self-peptides and microbial peptides that activated the AS- and AAU-derived TCRs. Structural analysis revealed that TCR cross-reactivity for peptide-MHC was rooted in a shared binding motif present in both self-antigens and microbial antigens that engages the BV9-CDR3β TCRs. These findings support the hypothesis that microbial antigens and self-antigens could play a pathogenic role in HLA-B*27-associated disease.
Organizational Affiliation:
Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, CA, USA.