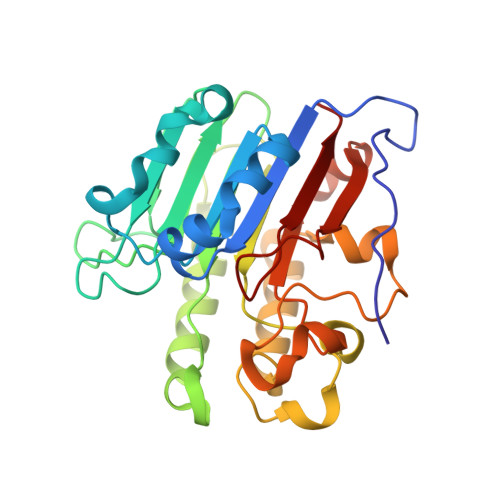
Capturing snapshots of APE1 processing DNA damage.
Freudenthal, B.D., Beard, W.A., Cuneo, M.J., Dyrkheeva, N.S., Wilson, S.H.(2015) Nat Struct Mol Biol 22: 924-931
- PubMed: 26458045
- DOI: https://doi.org/10.1038/nsmb.3105
- Primary Citation of Related Structures:
5DFF, 5DFH, 5DFI, 5DFJ, 5DG0 - PubMed Abstract:
DNA apurinic-apyrimidinic (AP) sites are prevalent noncoding threats to genomic stability and are processed by AP endonuclease 1 (APE1). APE1 incises the AP-site phosphodiester backbone, generating a DNA-repair intermediate that is potentially cytotoxic. The molecular events of the incision reaction remain elusive, owing in part to limited structural information. We report multiple high-resolution human APE1-DNA structures that divulge new features of the APE1 reaction, including the metal-binding site, the nucleophile and the arginine clamps that mediate product release. We also report APE1-DNA structures with a T-G mismatch 5' to the AP site, representing a clustered lesion occurring in methylated CpG dinucleotides. These structures reveal that APE1 molds the T-G mismatch into a unique Watson-Crick-like geometry that distorts the active site, thus reducing incision. These snapshots provide mechanistic clarity for APE1 while affording a rational framework to manipulate biological responses to DNA damage.
Organizational Affiliation:
Genome Integrity and Structural Biology Laboratory, National Institute of Environmental Health Sciences (NIEHS), National Institutes of Health, Research Triangle Park, USA.