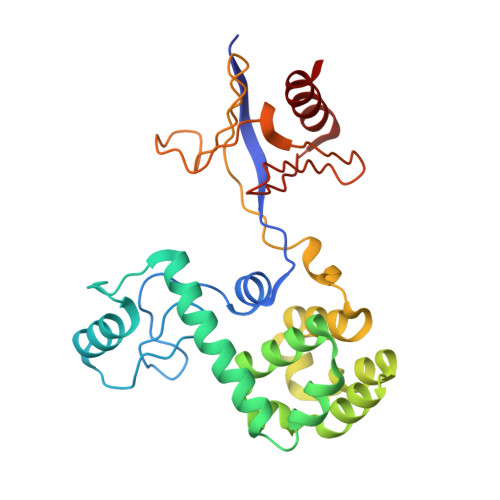
Structure of osh3 reveals a conserved mode of phosphoinositide binding in oxysterol-binding proteins
Tong, J., Yang, H., Yang, H., Eom, S.H., Im, Y.J.(2013) Structure 21: 1203-1213
- PubMed: 23791945
- DOI: https://doi.org/10.1016/j.str.2013.05.007
- Primary Citation of Related Structures:
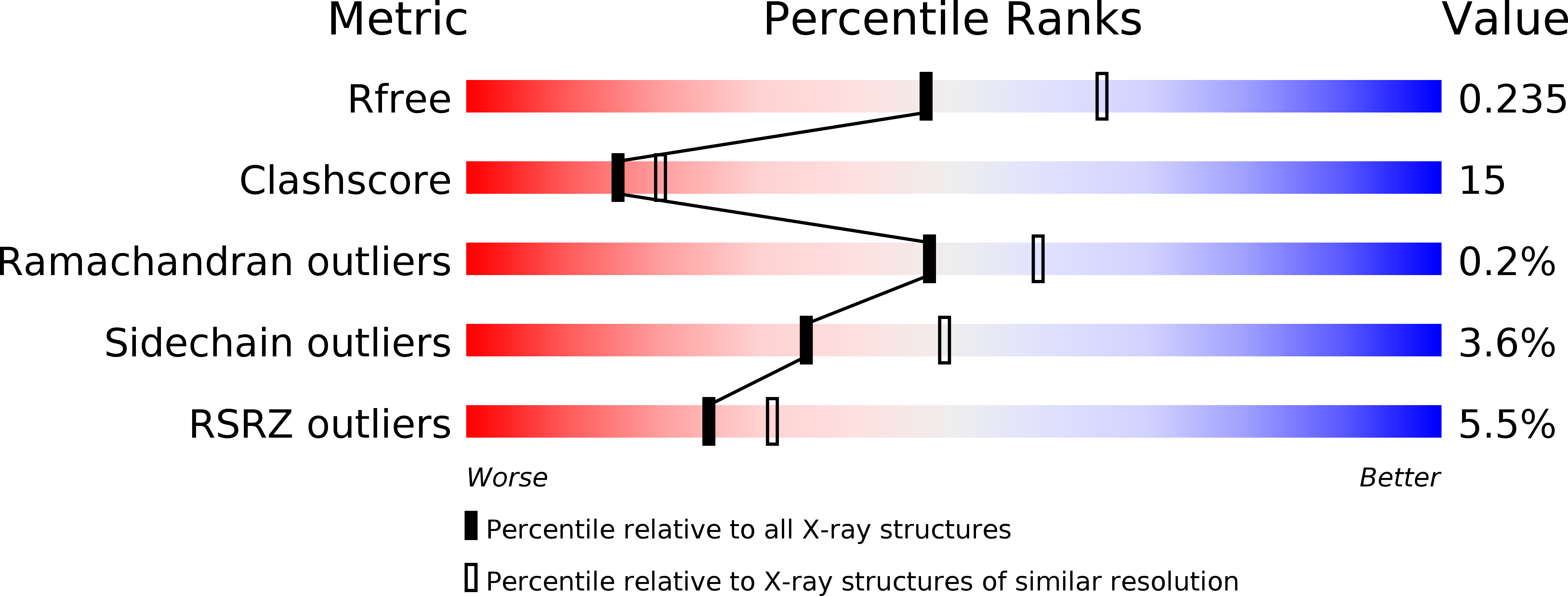
4IAP, 4IC4, 4INQ - PubMed Abstract:
The oxysterol-binding protein (OSBP)-related proteins (ORPs) are conserved from yeast to humans, and implicated in the regulation of lipid homeostasis and in signaling pathways. Saccharomyces cerevisiae has seven ORPs (Osh1-Osh7) that share one unknown essential function. Here, we report the 1.5-2.3 Å structures of the PH domain and ORD (OSBP-related domain) of yeast Osh3 in apo-form or in complex with phosphatidylinositol 4-phosphate (PI[4]P). Osh3 recognizes PI(4)P by the highly conserved residues in the tunnel of ORD whereas it lacks sterol binding due to the narrow hydrophobic tunnel. Yeast complementation tests suggest that PI(4)P binding to PH and ORD is essential for function. This study suggests that the unifying feature in all ORP homologs is the binding of PI(4)P to ORD and sterol binding is additional to certain homologs. Structural modeling of full-length Osh3 is consistent with the concept that Osh3 is a lipid transfer protein or regulator in membrane contact sites.
Organizational Affiliation:
College of Pharmacy, Chonnam National University, Gwangju 500-757, South Korea.