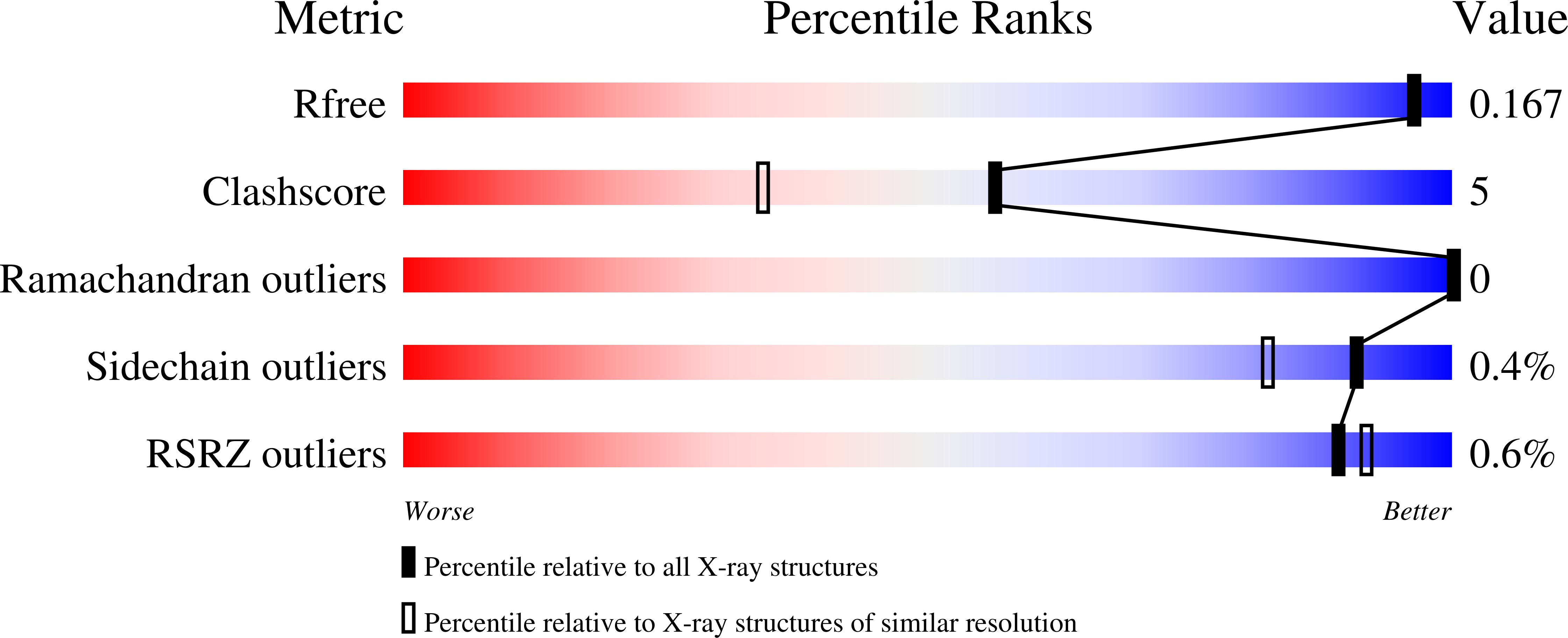
Dissecting the hydrophobic effect on the molecular level: the role of water, enthalpy, and entropy in ligand binding to thermolysin.
Biela, A., Nasief, N.N., Betz, M., Heine, A., Hangauer, D., Klebe, G.(2013) Angew Chem Int Ed Engl 52: 1822-1828
- PubMed: 23283700
- DOI: https://doi.org/10.1002/anie.201208561
- Primary Citation of Related Structures:
4H57
Organizational Affiliation:
Department of Pharmaceutical Chemistry, Philipps-University Marburg, Marbacher Weg 6, 35032 Marburg, Germany.