Purification, crystallization and preliminary X-ray diffraction studies on goat (Capra hircus) hemoglobin - a low oxygen affinity species
Sathya Moorthy, P., Neelagandan, K., Balasubramanian, M., Ponnuswamy, M.N.(2009) Protein Pept Lett 16: 454-456
- PubMed: 19356147
- DOI: https://doi.org/10.2174/092986609787847992
- Primary Citation of Related Structures:
3EU1 - PubMed Abstract:
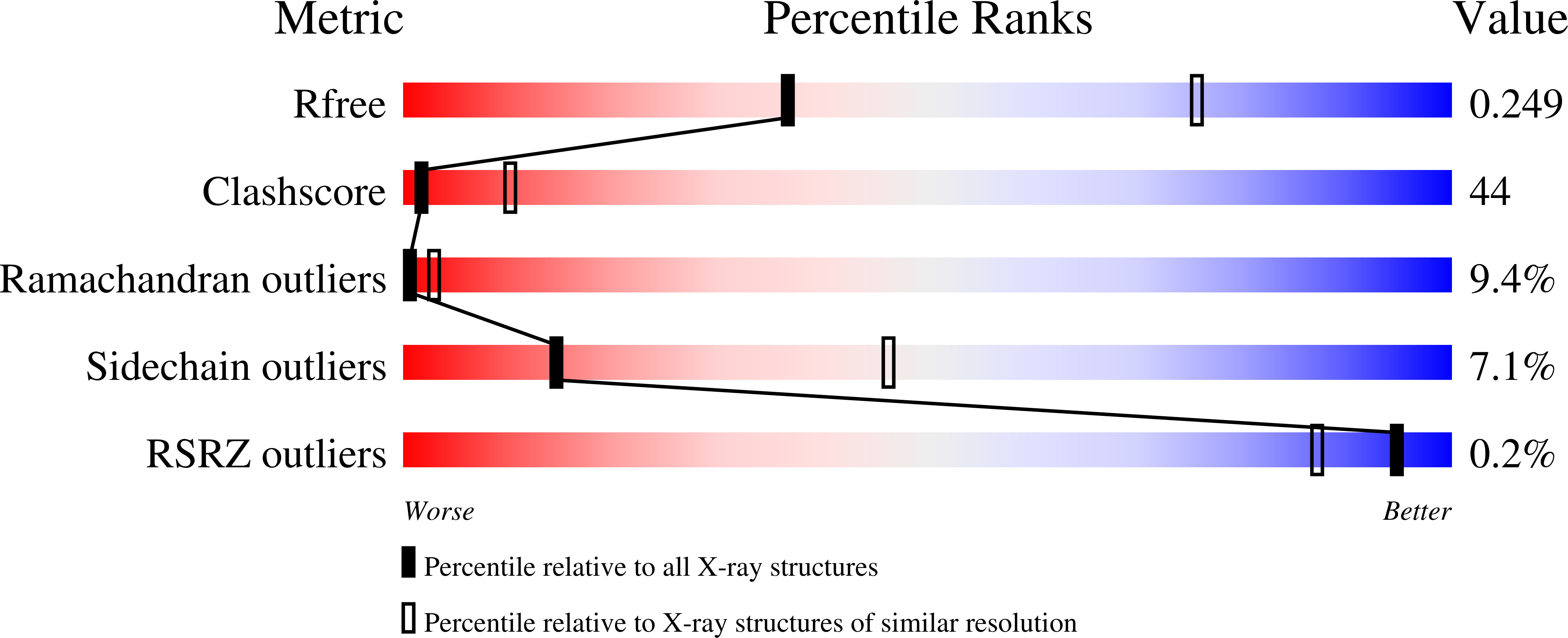
Hemoglobin is a vital protein present in almost all higher species. It is a transport protein involved in carrying oxygen from lungs to tissues and carbon dioxide back to lungs by an intrinsically coordinated manner. Even though a good amount of work has been carried out in this direction there exists scarcity of structural insight on low oxygen affinity species. Attempts are being made to unravel the structural insight of this low oxygen affinity species. Goat blood plasma was collected, treated with EDTA to avoid blood clotting and purification was accomplished using DEAE-anion chromatographic column. The goat hemoglobin was crystallized using 50mM of phosphate buffer at pH 6.7 with 1M NaCl and PEG 3350 as precipitant by hanging drop vapor diffusion method. Crystals obtained are screened and suitable crystals are taken for data collection using mar345dtb as image plate detector system. Goat hemoglobin crystal diffracted up to 2.61 A resolution. Goat hemoglobin crystallizes in orthorhombic space group P212(1)2(1) as a whole biological molecule in the asymmetric unit with cell dimensions a=53.568A, b=67.365A, c=154.183A.
Organizational Affiliation:
Centre of Advanced Study in Crystallography and Biophysics, University of Madras, Guindy Campus, Chennai - 600 025, India.