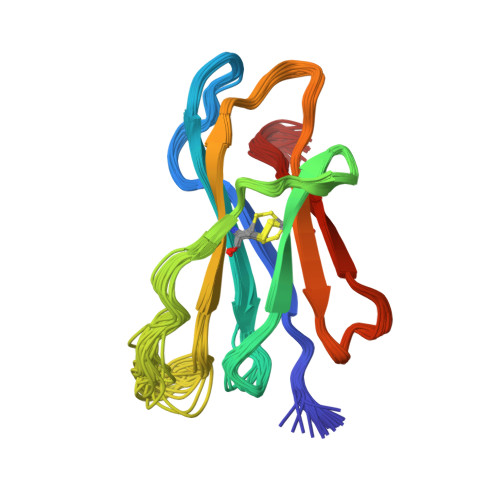
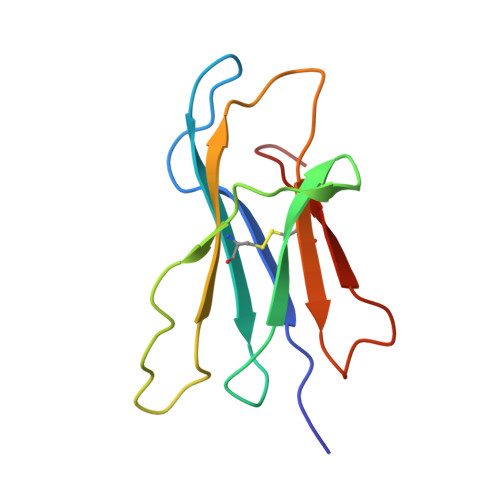
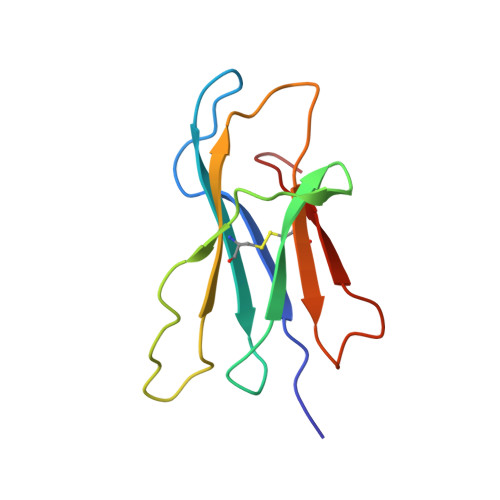
Conformational Conversion During Amyloid Formation at Atomic Resolution.
Eichner, T., Kalverda, A.P., Thompson, G.S., Homans, S.W., Radford, S.E.(2011) Mol Cell 41: 161-172
- PubMed: 21255727
- DOI: https://doi.org/10.1016/j.molcel.2010.11.028
- Primary Citation of Related Structures:
2XKS, 2XKU - PubMed Abstract:
Numerous studies of amyloid assembly have indicated that partially folded protein species are responsible for initiating aggregation. Despite their importance, the structural and dynamic features of amyloidogenic intermediates and the molecular details of how they cause aggregation remain elusive. Here, we use ΔN6, a truncation variant of the naturally amyloidogenic protein β(2)-microglobulin (β(2)m), to determine the solution structure of a nonnative amyloidogenic intermediate at high resolution. The structure of ΔN6 reveals a major repacking of the hydrophobic core to accommodate the nonnative peptidyl-prolyl trans-isomer at Pro32. These structural changes, together with a concomitant pH-dependent enhancement in backbone dynamics on a microsecond-millisecond timescale, give rise to a rare conformer with increased amyloidogenic potential. We further reveal that catalytic amounts of ΔN6 are competent to convert nonamyloidogenic human wild-type β(2)m (Hβ(2)m) into a rare amyloidogenic conformation and provide structural evidence for the mechanism by which this conformational conversion occurs.
Organizational Affiliation:
Astbury Centre for Structural Molecular Biology and Institute of Molecular and Cellular Biology, University of Leeds, Leeds LS2 9JT, UK.