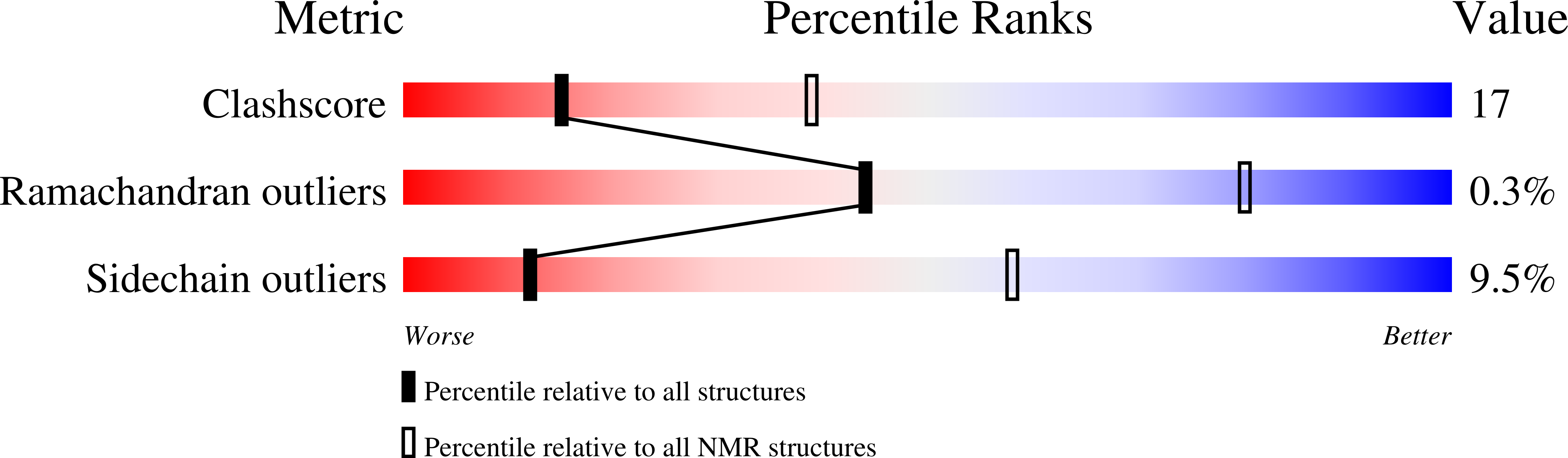The dilated cardiomyopathy G159D mutation in cardiac troponin C weakens the anchoring interaction with troponin I.
Baryshnikova, O.K., Robertson, I.M., Mercier, P., Sykes, B.D.(2008) Biochemistry 47: 10950-10960
- PubMed: 18803402
- DOI: https://doi.org/10.1021/bi801165c
- Primary Citation of Related Structures:
2MLE, 2MLF - PubMed Abstract:
NMR spectroscopy has been employed to elucidate the molecular consequences of the DCM G159D mutation on the structure and dynamics of troponin C, and its interaction with troponin I (TnI). Since the molecular effects of human mutations are often subtle, all NMR experiments were conducted as direct side-by-side comparisons of the wild-type C-domain of troponin C (cCTnC) and the mutant protein, G159D. With the mutation, the affinity toward the anchoring region of cTnI (cTnI 34-71) was reduced ( K D = 3.0 +/- 0.6 microM) compared to that of the wild type ( K D < 1 microM). Overall, the structure and dynamics of the G159D.cTnI 34-71 complex were very similar to those of the cCTnC.cTnI 34-71 complex. There were, however, significant changes in the (1)H, (13)C, and (15)N NMR chemical shifts, especially for the residues in direct contact with cTnI 34-71, and the changes in NOE connectivity patterns between the G159D.cTnI 34-71 and cCTnC.cTnI 34-71 complexes. Thus, the most parsimonious hypothesis is that the development of disease results from the poor anchoring of cTnI to cCTnC, with the resulting increase in the level of acto-myosin inhibition in agreement with physiological data. Another possibility is that long-range electrostatic interactions affect the binding of the inhibitory and switch regions of cTnI (cTnI 128-147 and cTnI 147-163) and/or the cardiac specific N-terminus of cTnI (cTnI 1-29) to the N-domain of cTnC. These important interactions are all spatially close in the X-ray structure of the cardiac TnC core.
Organizational Affiliation:
Department of Biochemistry, University of Alberta, Edmonton, Alberta, Canada.