Prion Protein mPrP[F175A](121-231): Structure and Stability in Solution.
Christen, B., Hornemann, S., Damberger, F.F., Wuthrich, K.(2012) J Mol Biol 423: 496-502
- PubMed: 22922482
- DOI: https://doi.org/10.1016/j.jmb.2012.08.011
- Primary Citation of Related Structures:
2L1E - PubMed Abstract:
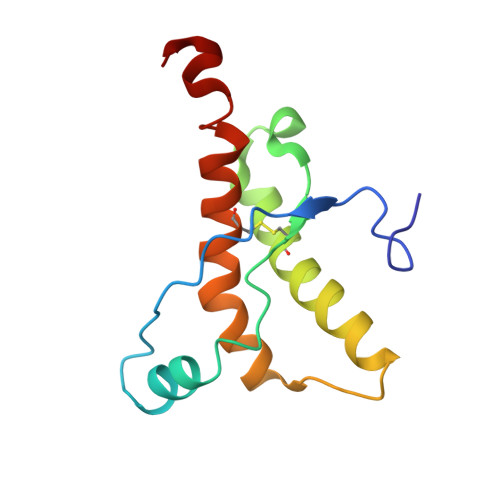
The three-dimensional structures of prion proteins (PrPs) in the cellular form (PrP(C)) include a stacking interaction between the aromatic rings of the residues Y169 and F175, where F175 is conserved in all but two so far analyzed mammalian PrP sequences and where Y169 is strictly conserved. To investigate the structural role of F175, we characterized the variant mouse prion protein mPrP[F175A](121-231). The NMR solution structure represents a typical PrP(C)-fold, and it contains a 3(10)-helical β2-α2 loop conformation, which is well defined because all amide group signals in this loop are observed at 20°C. With this "rigid-loop PrP(C)" behavior, mPrP[F175A](121-231) differs from the previously studied mPrP[Y169A](121-231), which contains a type I β-turn β2-α2 loop structure. When compared to other rigid-loop variants of mPrP(121-231), mPrP[F175A](121-231) is unique in that the thermal unfolding temperature is lowered by 8°C. These observations enable further refined dissection of the effects of different single-residue exchanges on the PrP(C) conformation and their implications for the PrP(C) physiological function.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, ETH Zurich, Schafmattstrasse 20, CH-8093 Zurich, Switzerland.