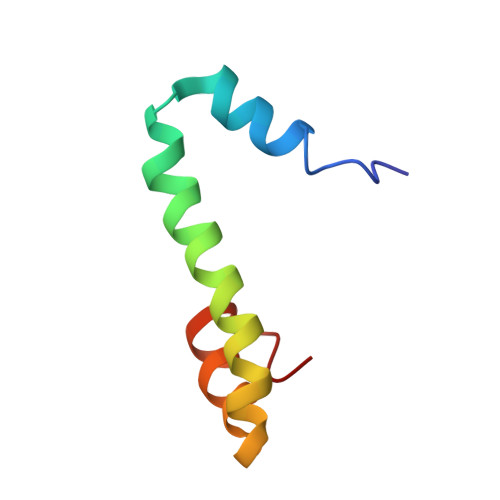
Nogo goes in the pure water: solution structure of Nogo-60 and design of the structured and buffer-soluble Nogo-54 for enhancing CNS regeneration
Li, M.F., Liu, J.X., Song, J.X.(2006) Protein Sci 15: 1835-1841
- PubMed: 16877707
- DOI: https://doi.org/10.1110/ps.062306906
- Primary Citation of Related Structures:
2G31 - PubMed Abstract:
The inability to determine the structure of the buffer-insoluble Nogo extracellular domain retarded further design of Nogo receptor (NgR) antagonists to treat CNS axonal injuries. Very surprisingly, we recently discovered that Nogo-60 was soluble and structured in salt-free water, thus allowing the determination of the first Nogo structure by heteronuclear NMR spectroscopy. Nogo-60 adopts an unusual helical structure with the N- and C-terminal helices connected by a long middle helix. While the N-helix has no contact with the rest of the molecule, the C-helix flips back to pack against the 20-residue middle helix. This packing appears to trigger the formation of the stable Nogo-60 structure because Nogo-40 with the last helix truncated is unstructured. The Nogo-60 structure offered us rationales for further design of the structured and buffer-soluble Nogo-54, which may be used as a novel NgR antagonist. Furthermore, our discovery may imply a general solution to solubilizing a category of buffer-insoluble proteins for urgent structural investigations.
Organizational Affiliation:
Department of Biological Sciences, Faculty of Science, National University of Singapore, Singapore.