Solution structure of the Rhodobacter sphaeroides PufX membrane protein: implications for the quinone exchange and protein-protein interactions
Wang, Z.Y., Suzuki, H., Kobayashi, M., Nozawa, T.(2007) Biochemistry 46: 3635-3642
- PubMed: 17335288
- DOI: https://doi.org/10.1021/bi0618060
- Primary Citation of Related Structures:
2DW3 - PubMed Abstract:
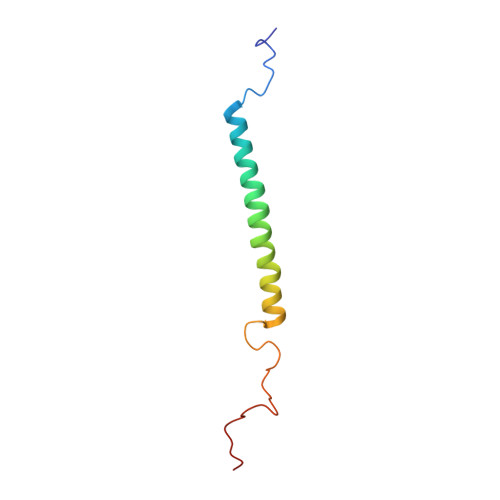
PufX membrane protein is found in Rhodobacter species of purple photosynthetic bacteria and has been known to play an essential role in ubiquinone/ubiquinol exchange between the reaction center and cytochrome bc1 complex and also contribute to the dimerization of the reaction center-light-harvesting core complex. We have determined the solution structure of the Rhodobacter sphaeroides PufX using multidimensional NMR spectroscopy. The PufX, functionally expressed in Escherichia coli, forms a stable alpha helix consisting of 21 residues over the central transmembrane domain. The overall structure of the PufX is very similar to those of the LH1 alpha- and beta-polypeptides from Rhodospirillum rubrum and LH2 polypeptides. A short segment (Lys28-Gly35) rich in Gly and Ala residues revealed a relatively fast exchange between the backbone amide protons and deuteriums in the hydroxyl groups of the solvent, indicating that the backbone of this segment is more easily accessible to the surrounding solvent molecules compared to those of its neighboring portions. The Gly- and Ala-rich segment is located in the middle of the central helix and forms an extensive groove-like conformation on the surface with the neighboring residues, where the residues with large side chains are aligned on one side of the helix, and small residues are aligned on the other face. Such a structural motif may serve as a functional site responsible for ubiquinol transport from the core complex to the membrane phase and for sequence-specific helix-helix interactions with the neighboring polypeptides.
Organizational Affiliation:
Faculty of Science, Ibaraki University, Mito 310-8512, Japan. wang@mx.ibaraki.ac.jp