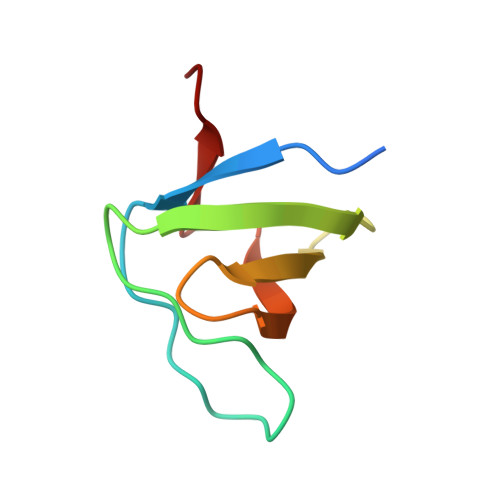
Structural Investigation of the Binding of a Herpesviral Protein to the SH3 Domain of Tyrosine Kinase Lck.
Schweimer, K., Hoffmann, S., Bauer, F., Friedrich, U., Kardinal, C., Feller, S.M., Biesinger, B., Sticht, H.(2002) Biochemistry 41: 5120
- PubMed: 11955060
- DOI: https://doi.org/10.1021/bi015986j
- Primary Citation of Related Structures:
1H92, 1WA7 - PubMed Abstract:
Herpesvirus saimiri codes for a tyrosine kinase interacting protein (Tip) that interacts with both the SH3 domain and the kinase domain of the T-cell-specific tyrosine kinase Lck via two separate motifs. The activation of Lck by Tip is considered as a key event in the transformation of human T-lymphocytes during herpesviral infection. We investigated the interaction of proline-rich Tip peptides with the LckSH3 domain starting with the structural characterization of the unbound interaction partners. The solution structure of the LckSH3 was determined by heteronuclear multidimensional nuclear magnetic resonance (NMR) spectroscopy using 44 residual dipolar couplings in addition to the conventional experimental restraints. Circular dichroism spectroscopy proved that the polyproline helix of Tip is already formed prior to SH3 binding and is conformationally stable. NMR titration experiments point out three major regions of the Tip-Lck interaction comprising the RT loop, the n-src loop, and a helical turn preceding the last strand of the beta-sheet. Further changes of the chemical shifts were observed for the N- and C-terminal beta-strands of the SH3 domain, indicating additional contacts outside the proline-rich segment or subtle structural rearrangements transmitted from the binding site of the proline helix. Fluorescence spectroscopy shows that Tip binds to the SH3 domains of several Src kinases (Lck, Hck, Lyn, Src, Fyn, Yes), exhibiting the highest affinities for Lyn, Hck, and Lck.
Organizational Affiliation:
Lehrstuhl für Biopolymere, Universität Bayreuth, 95440 Bayreuth, Germany.