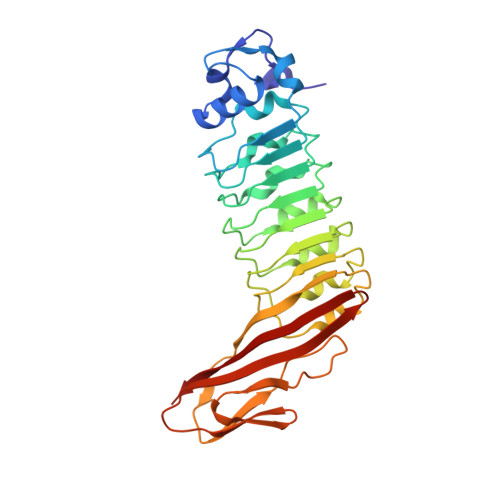
Internalins from the human pathogen Listeria monocytogenes combine three distinct folds into a contiguous internalin domain.
Schubert, W.D., Gobel, G., Diepholz, M., Darji, A., Kloer, D., Hain, T., Chakraborty, T., Wehland, J., Domann, E., Heinz, D.W.(2001) J Mol Biol 312: 783-794
- PubMed: 11575932
- DOI: https://doi.org/10.1006/jmbi.2001.4989
- Primary Citation of Related Structures:
1H6T, 1H6U - PubMed Abstract:
Listeria monocytogenes is an opportunistic, food-borne human and animal pathogen. Host cell invasion requires the action of the internalins A (InlA) and B (InlB), which are members of a family of listerial cell-surface proteins. Common to these proteins are three distinctive N-terminal domains that have been shown to direct host cell-specific invasion for InlA and InlB. Here, we present the high-resolution crystal structures of these domains present in InlB and InlH, and show that they constitute a single "internalin domain". In this internalin domain, a central LRR region is flanked contiguously by a truncated EF-hand-like cap and an immunoglobulin (Ig)-like fold. The extended beta-sheet, resulting from the distinctive fusion of the LRR and the Ig-like folds, constitutes an adaptable concave interaction surface, which we propose is responsible for the specific recognition of the host cellular binding partners during infection.
Organizational Affiliation:
Department of Structural Biology, German Research Center for Biotechnology, Braunschweig.