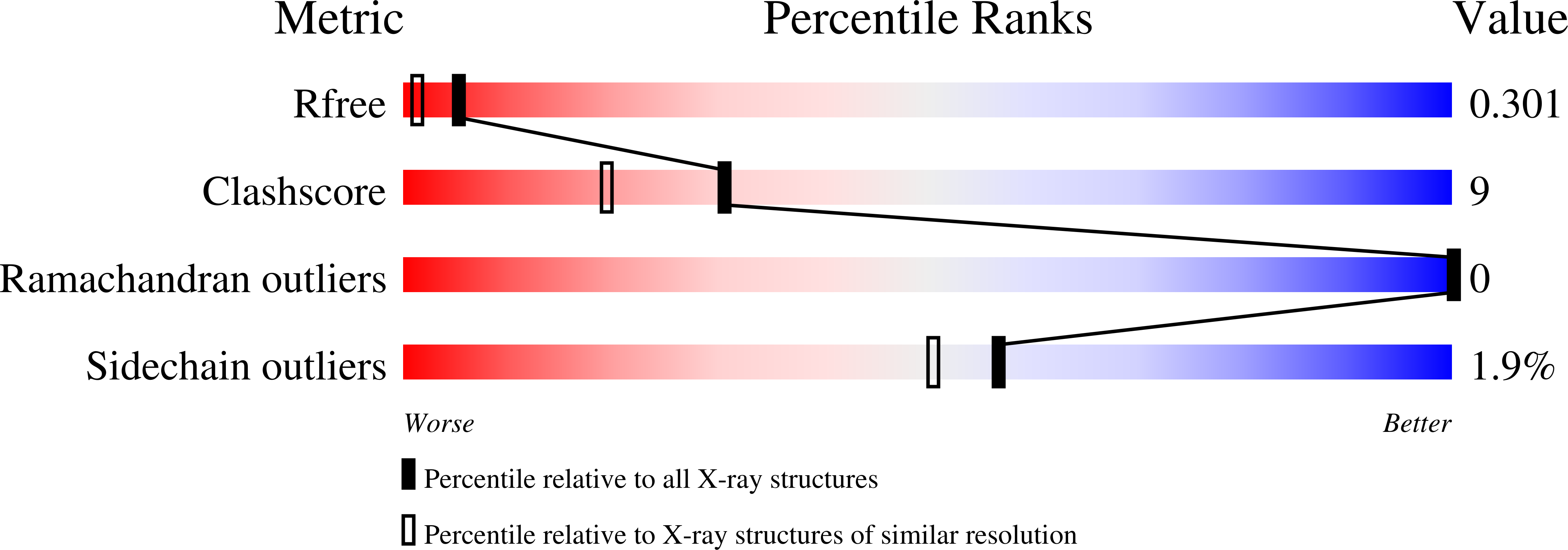
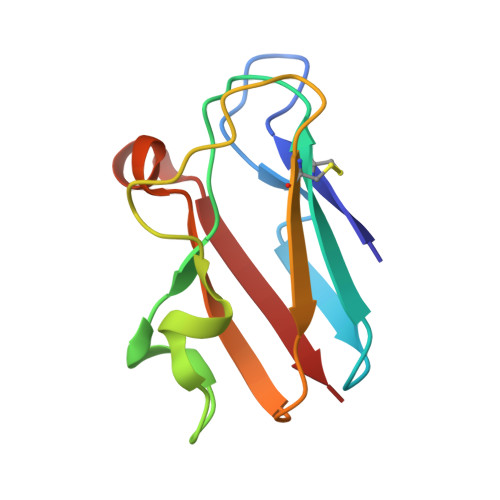
Novel Disulfide Bonds Effect the Thermostability of Plastocyanin. Crystal structures of the triple plastocyanin mutant G8D/K30C/T69C and the double plastocyanin mutant K30C/T69C from spinach at 1.90 A and 1.96 A resolution, respectively.
Okvist, M., Jacobson, F., Jansson, H., Hansson, O., Sjolin, L.To be published.