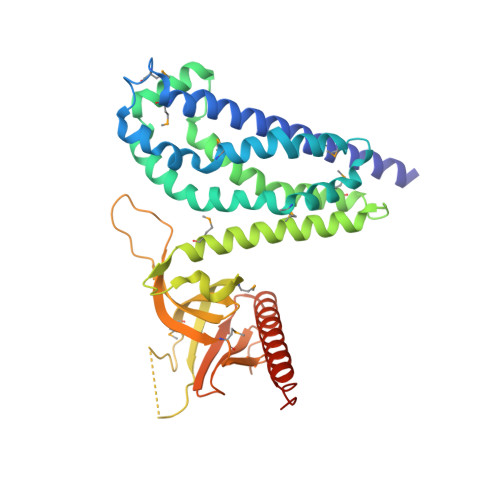
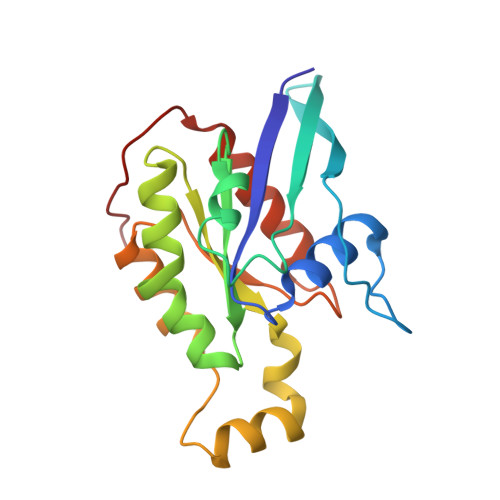
A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange.
Rossman, K.L., Worthylake, D.K., Snyder, J.T., Siderovski, D.P., Campbell, S.L., Sondek, J.(2002) EMBO J 21: 1315-1326
- PubMed: 11889037
- DOI: https://doi.org/10.1093/emboj/21.6.1315
- Primary Citation of Related Structures:
1KZ7, 1KZG - PubMed Abstract:
Dbl-related oncoproteins are guanine nucleotide exchange factors (GEFs) specific for Rho guanosine triphosphatases (GTPases) and invariably possess tandem Dbl (DH) and pleckstrin homology (PH) domains. While it is known that the DH domain is the principal catalytic subunit, recent biochemical data indicate that for some Dbl-family proteins, such as Dbs and Trio, PH domains may cooperate with their associated DH domains in promoting guanine nucleotide exchange of Rho GTPases. In order to gain an understanding of the involvement of these PH domains in guanine nucleotide exchange, we have determined the crystal structure of a DH/PH fragment from Dbs in complex with Cdc42. The complex features the PH domain in a unique conformation distinct from the PH domains in the related structures of Sos1 and Tiam1.Rac1. Consequently, the Dbs PH domain participates with the DH domain in binding Cdc42, primarily through a set of interactions involving switch 2 of the GTPase. Comparative sequence analysis suggests that a subset of Dbl-family proteins will utilize their PH domains similarly to Dbs.
Organizational Affiliation:
Department of Biochemistry and Biophysics, University of North Carolina, Chapel Hill, NC 27599, USA.