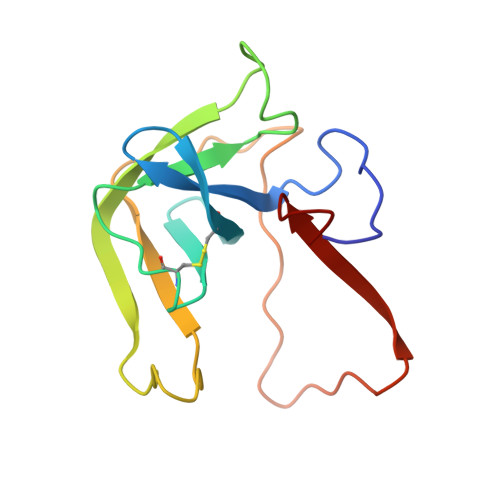
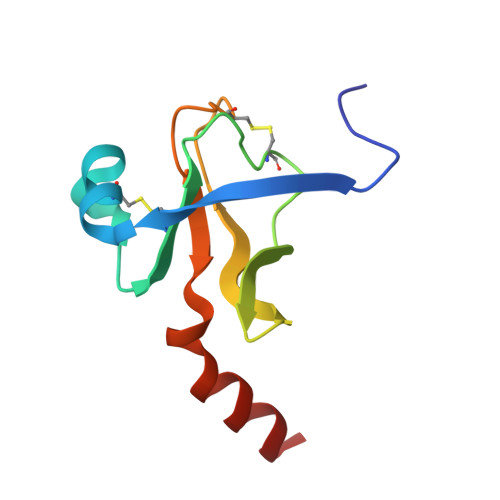
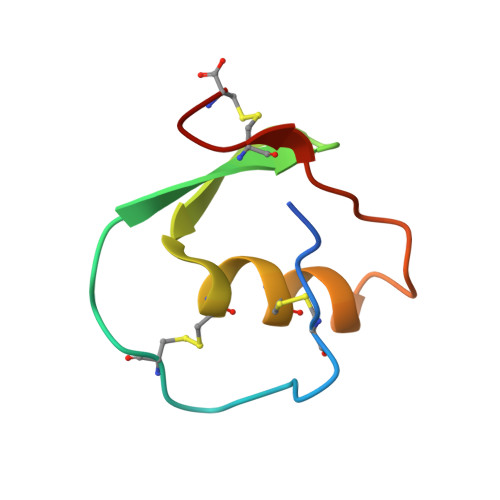
Crystal and molecular structures of the complex of alpha-chymotrypsin with its inhibitor turkey ovomucoid third domain at 1.8 A resolution.
Fujinaga, M., Sielecki, A.R., Read, R.J., Ardelt, W., Laskowski Jr., M., James, M.N.(1987) J Mol Biol 195: 397-418
- PubMed: 3477645
- DOI: https://doi.org/10.1016/0022-2836(87)90659-0
- Primary Citation of Related Structures:
1CHO - PubMed Abstract:
The molecular structure of the complex between bovine pancreatic alpha-chymotrypsin (EC 3.4.4.5) and the third domain of the Kazal-type ovomucoid from Turkey (OMTKY3) has been determined crystallographically by the molecular replacement method. Restrained-parameter least-squares refinement of the molecular model of the complex has led to a conventional agreement factor R of 0.168 for the 19,466 reflections in the 1.8 A (1 A = 0.1 nm) resolution shell [I greater than or equal to sigma (I)]. The reactive site loop of OMTKY3, from Lys13I to Arg21I (I indicates inhibitor), is highly complementary to the surface of alpha-chymotrypsin in the complex. A total of 13 residues on the inhibitor make 113 contacts of less than 4.0 A with 21 residues of the enzyme. A short contact (2.95 A) from O gamma of Ser195 to the carbonyl-carbon atom of the scissile bond between Leu18I and Glu19I is present; in spite of it, this peptide remains planar and undistorted. Analysis of the interactions of the inhibitor with chymotrypsin explains the enhanced specificity that chymotrypsin has for P'3 arginine residues. There is a water-mediated ion pair between the guanidinium group on this residue and the carboxylate of Asp64. Comparison of the structure of the alpha-chymotrypsin portion of this complex with the several structures of alpha and gamma-chymotrypsin in the uncomplexed form shows a high degree of structural equivalence (root-mean-square deviation of the 234 common alpha-carbon atoms averages 0.38 A). Significant differences occur mainly in two regions Lys36 to Phe39 and Ser75 to Lys79. Among the 21 residues that are in contact with the ovomucoid domain, only Phe39 and Tyr146 change their conformations significantly as a result of forming the complex. Comparison of the structure of the OMTKY3 domain in this complex to that of the same inhibitor bound to a serine proteinase from Streptomyces griseus (SGPB) shows a central core of 44 amino acids (the central alpha-helix and flanking small 3-stranded beta-sheet) that have alpha-carbon atoms fitting to within 1.0 A (root-mean-square deviation of 0.45 A) whereas the residues of the reactive-site loop differ in position by up to 1.9 A (C alpha of Leu18I). The ovomucoid domain has a built-in conformational flexibility that allows it to adapt to the active sites of different enzymes. A comparison of the SGPB and alpha-chymotrypsin molecules is made and the water molecules bound at the inhibitor-enzyme interface in both complexes are analysed for similarities and differences.
Organizational Affiliation:
Department of Biochemistry, University of Alberta, Edmonton, Canada.