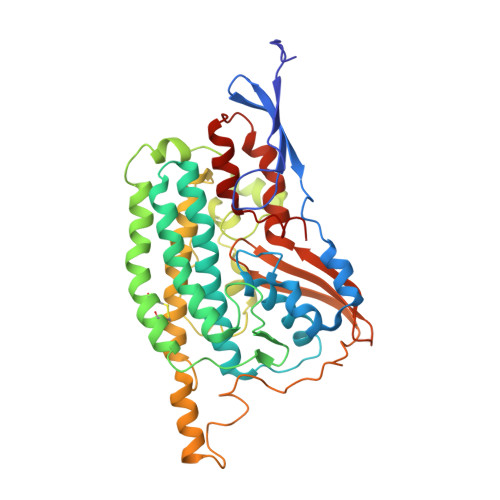
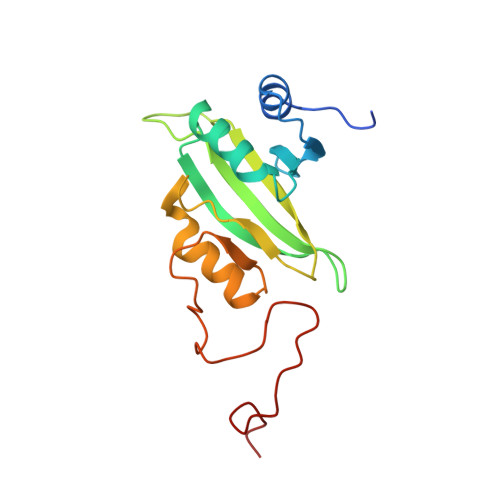
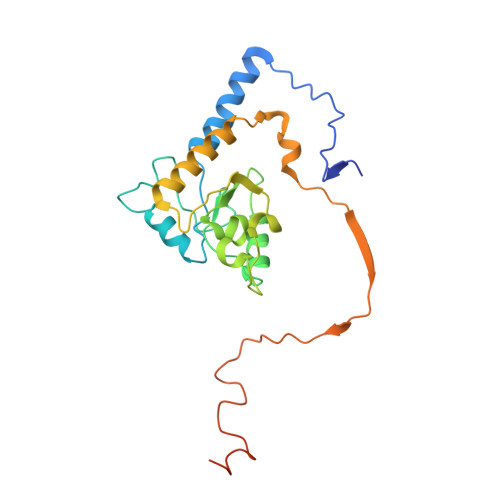
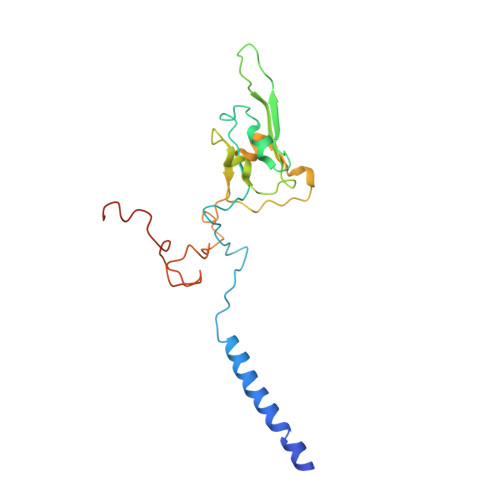
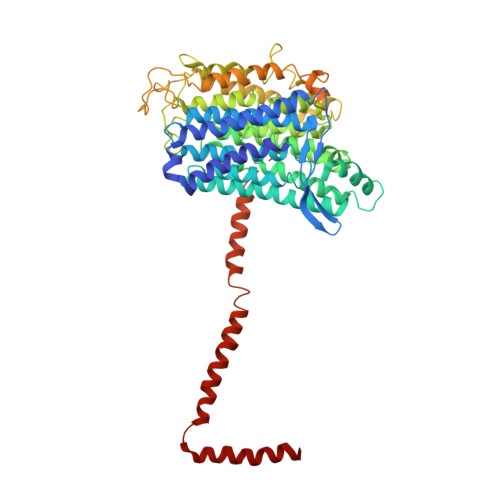
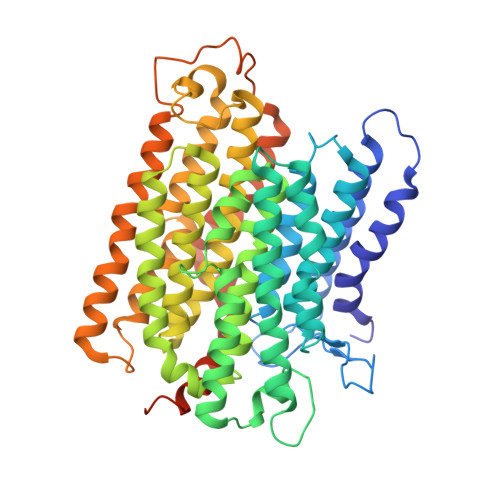
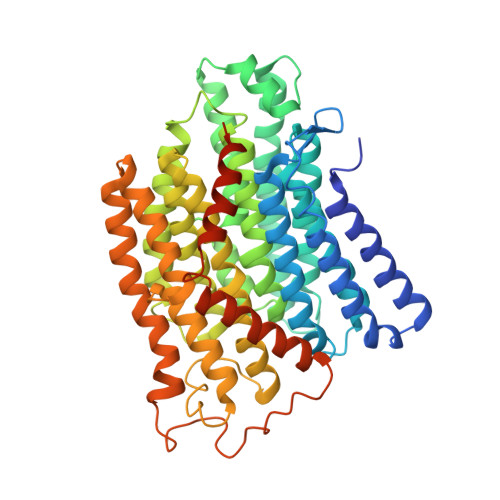
Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Laughlin, T.G., Bayne, A.N., Trempe, J.F., Savage, D.F., Davies, K.M.(2019) Nature 566: 411-414
- PubMed: 30742075
- DOI: https://doi.org/10.1038/s41586-019-0921-0
- PubMed Abstract:
Cyclic electron flow around photosystem I (PSI) is a mechanism by which photosynthetic organisms balance the levels of ATP and NADPH necessary for efficient photosynthesis 1,2 . NAD(P)H dehydrogenase-like complex (NDH) is a key component of this pathway in most oxygenic photosynthetic organisms 3,4 and is the last large photosynthetic membrane-protein complex for which the structure remains unknown. Related to the respiratory NADH dehydrogenase complex (complex I), NDH transfers electrons originating from PSI to the plastoquinone pool while pumping protons across the thylakoid membrane, thereby increasing the amount of ATP produced per NADP + molecule reduced 4,5 . NDH possesses 11 of the 14 core complex I subunits, as well as several oxygenic-photosynthesis-specific (OPS) subunits that are conserved from cyanobacteria to plants 3,6 . However, the three core complex I subunits that are involved in accepting electrons from NAD(P)H are notably absent in NDH 3,5,6 , and it is therefore not clear how NDH acquires and transfers electrons to plastoquinone. It is proposed that the OPS subunits-specifically NdhS-enable NDH to accept electrons from its electron donor, ferredoxin 3-5,7 . Here we report a 3.1 Å structure of the 0.42-MDa NDH complex from the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1, obtained by single-particle cryo-electron microscopy. Our maps reveal the structure and arrangement of the principal OPS subunits in the NDH complex, as well as an unexpected cofactor close to the plastoquinone-binding site in the peripheral arm. The location of the OPS subunits supports a role in electron transfer and defines two potential ferredoxin-binding sites at the apex of the peripheral arm. These results suggest that NDH could possess several electron transfer routes, which would serve to maximize plastoquinone reduction and avoid deleterious off-target chemistry of the semi-plastoquinone radical.
Organizational Affiliation:
Department of Molecular and Cell Biology, University of California, Berkeley, CA, USA.