Mechanisms of redundancy and specificity of the Aspergillus fumigatus Crh transglycosylases.
Fang, W., Sanz, A.B., Bartual, S.G., Wang, B., Ferenbach, A.T., Farkas, V., Hurtado-Guerrero, R., Arroyo, J., van Aalten, D.M.F.(2019) Nat Commun 10: 1669-1669
- PubMed: 30971696
- DOI: https://doi.org/10.1038/s41467-019-09674-0
- Primary Citation of Related Structures:
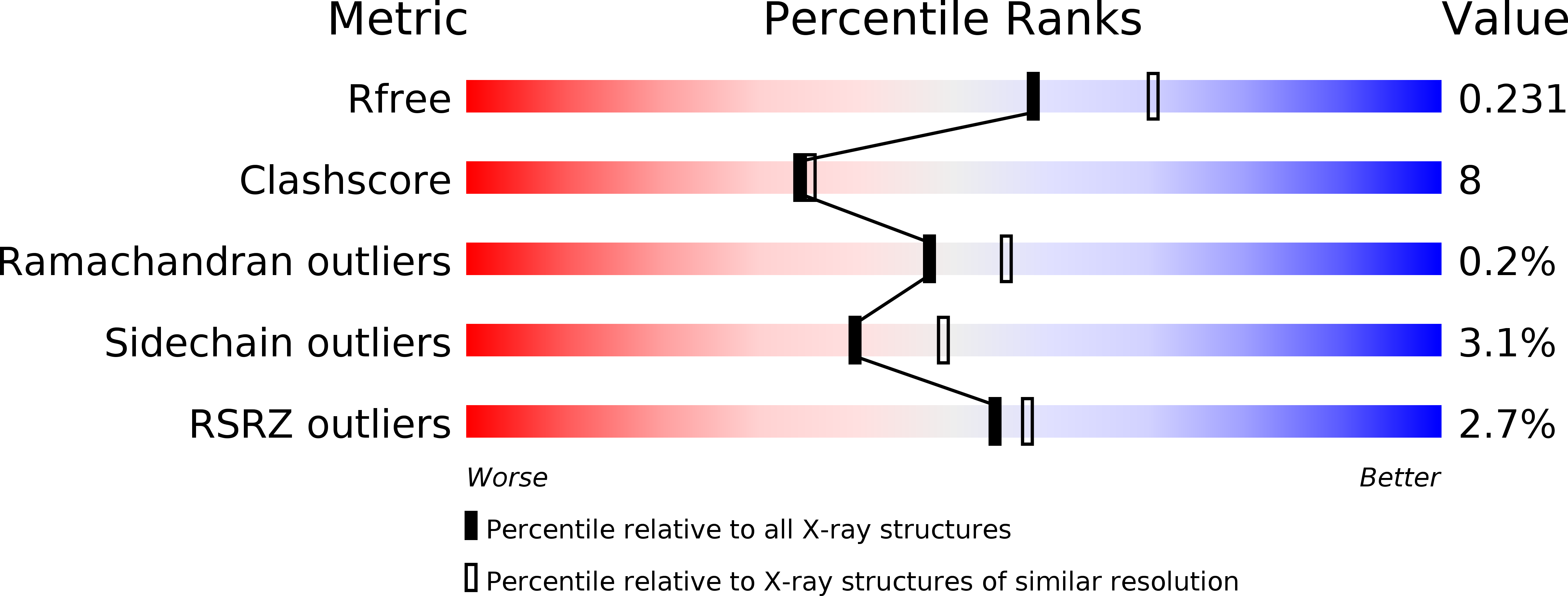
6IBU, 6IBW - PubMed Abstract:
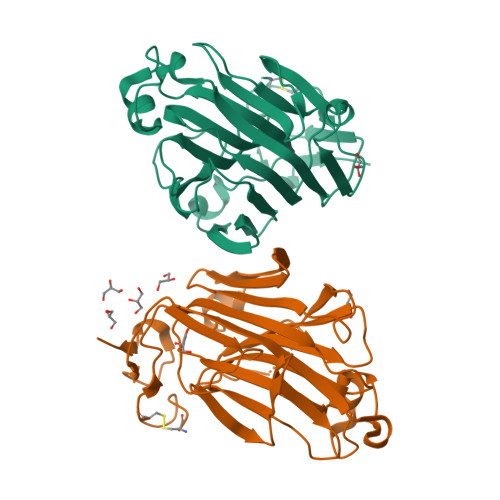
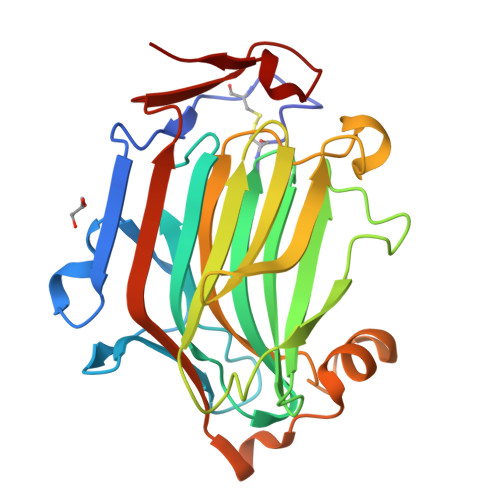
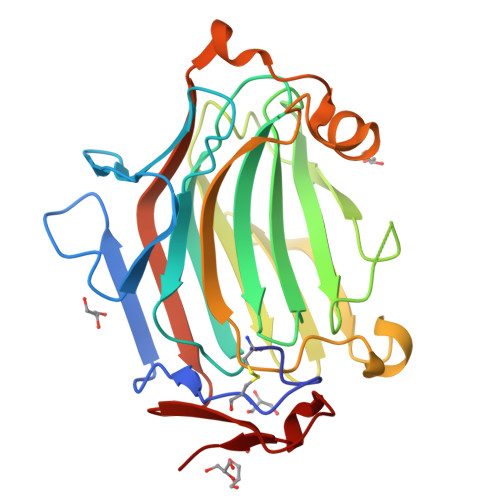
Fungal cell wall synthesis is achieved by a balance of glycosyltransferase, hydrolase and transglycosylase activities. Transglycosylases strengthen the cell wall by forming a rigid network of crosslinks through mechanisms that remain to be explored. Here we study the function of the Aspergillus fumigatus family of five Crh transglycosylases. Although crh genes are dispensable for cell viability, simultaneous deletion of all genes renders cells sensitive to cell wall interfering compounds. In vitro biochemical assays and localisation studies demonstrate that this family of enzymes functions redundantly as transglycosylases for both chitin-glucan and chitin-chitin cell wall crosslinks. To understand the molecular basis of this acceptor promiscuity, we solved the crystal structure of A. fumigatus Crh5 (AfCrh5) in complex with a chitooligosaccharide at the resolution of 2.8 Å, revealing an extensive elongated binding cleft for the donor (-4 to -1) substrate and a short acceptor (+1 to +2) binding site. Together with mutagenesis, the structure suggests a "hydrolysis product assisted" molecular mechanism favouring transglycosylation over hydrolysis.
Organizational Affiliation:
School of Life Sciences, University of Dundee, Dundee, DD1 5EH, UK.