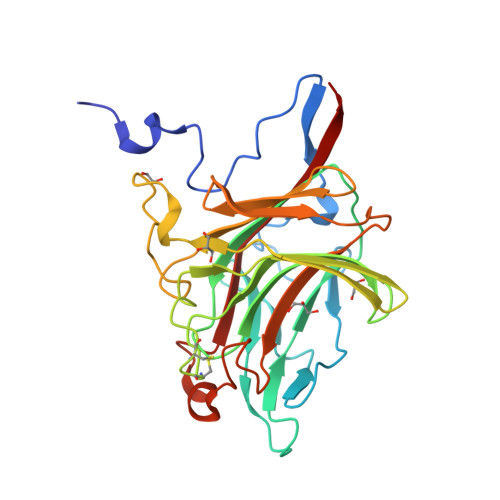
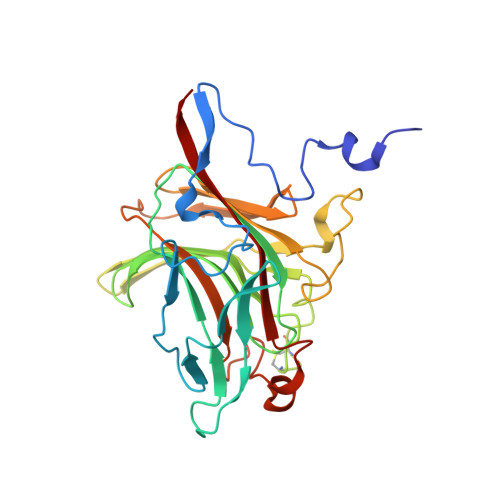
The agar-specific hydrolaseZgAgaC from the marine bacteriumZobellia galactanivoransdefines a new GH16 protein subfamily.
Naretto, A., Fanuel, M., Ropartz, D., Rogniaux, H., Larocque, R., Czjzek, M., Tellier, C., Michel, G.(2019) J Biological Chem 294: 6923-6939
- PubMed: 30846563
- DOI: https://doi.org/10.1074/jbc.RA118.006609
- Primary Citation of Related Structures:
6HY3 - PubMed Abstract:
Agars are sulfated galactans from red macroalgae and are composed of a d-galactose (G unit) and l-galactose (L unit) alternatively linked by α-1,3 and β-1,4 glycosidic bonds. These polysaccharides display high complexity, with numerous modifications of their backbone ( e.g. presence of a 3,6-anhydro-bridge (LA unit) and sulfations and methylation). Currently, bacterial polysaccharidases that hydrolyze agars (β-agarases and β-porphyranases) have been characterized on simple agarose and more rarely on porphyran, a polymer containing both agarobiose (G-LA) and porphyranobiose (GL6S) motifs. How bacteria can degrade complex agars remains therefore an open question. Here, we studied an enzyme from the marine bacterium Zobellia galactanivorans ( Zg AgaC) that is distantly related to the glycoside hydrolase 16 (GH16) family β-agarases and β-porphyranases. Using a large red algae collection, we demonstrate that Zg AgaC hydrolyzes not only agarose but also complex agars from Ceramiale s species. Using tandem MS analysis, we elucidated the structure of a purified hexasaccharide product, L6S-G-LA2Me-G(2Pentose)-LA2S-G, released by the activity of Zg AgaC on agar extracted from Osmundea pinnatifida By resolving the crystal structure of Zg AgaC at high resolution (1.3 Å) and comparison with the structures of Zg AgaB and Zg PorA in complex with their respective substrates, we determined that Zg AgaC recognizes agarose via a mechanism different from that of classical β-agarases. Moreover, we identified conserved residues involved in the binding of complex oligoagars and demonstrate a probable influence of the acidic polysaccharide's pH microenvironment on hydrolase activity. Finally, a phylogenetic analysis supported the notion that Zg AgaC homologs define a new GH16 subfamily distinct from β-porphyranases and classical β-agarases.
Organizational Affiliation:
From Sorbonne Université, CNRS, Integrative Biology of Marine Models (LBI2M), Station Biologique de Roscoff (SBR), 29680 Roscoff, Bretagne, France.