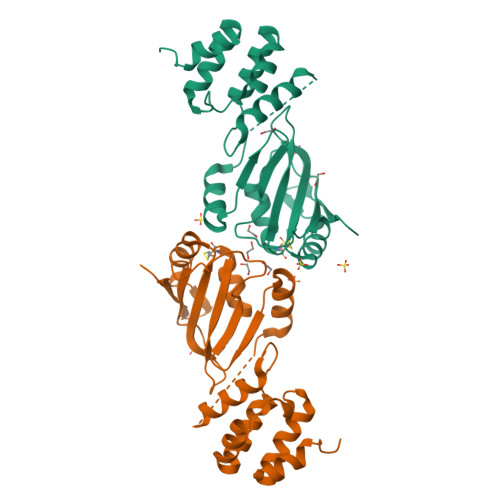
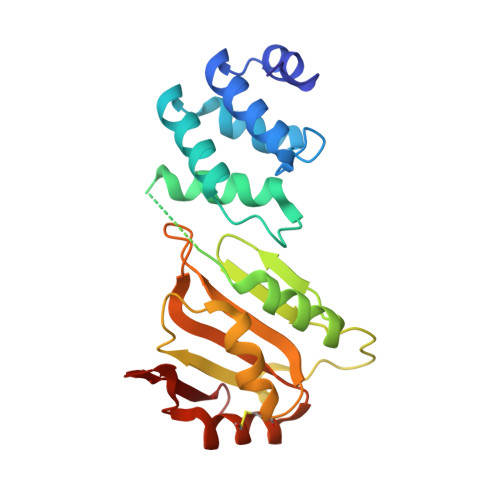
A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets.
Hobor, F., Dallmann, A., Ball, N.J., Cicchini, C., Battistelli, C., Ogrodowicz, R.W., Christodoulou, E., Martin, S.R., Castello, A., Tripodi, M., Taylor, I.A., Ramos, A.(2018) Nat Commun 9: 831-831
- PubMed: 29483512
- DOI: https://doi.org/10.1038/s41467-018-03182-3
- Primary Citation of Related Structures:
6ES4 - PubMed Abstract:
Exosomal miRNA transfer is a mechanism for cell-cell communication that is important in the immune response, in the functioning of the nervous system and in cancer. Syncrip/hnRNPQ is a highly conserved RNA-binding protein that mediates the exosomal partition of a set of miRNAs. Here, we report that Syncrip's amino-terminal domain, which was previously thought to mediate protein-protein interactions, is a cryptic, conserved and sequence-specific RNA-binding domain, designated NURR (N-terminal unit for RNA recognition). The NURR domain mediates the specific recognition of a short hEXO sequence defining Syncrip exosomal miRNA targets, and is coupled by a non-canonical structural element to Syncrip's RRM domains to achieve high-affinity miRNA binding. As a consequence, Syncrip-mediated selection of the target miRNAs implies both recognition of the hEXO sequence by the NURR domain and binding of the RRM domains 5' to this sequence. This structural arrangement enables Syncrip-mediated selection of miRNAs with different seed sequences.
Organizational Affiliation:
Research Department of Structural and Molecular Biology, University College London, Darwin Building, Gower Street, London, WC1E 6XA, UK.