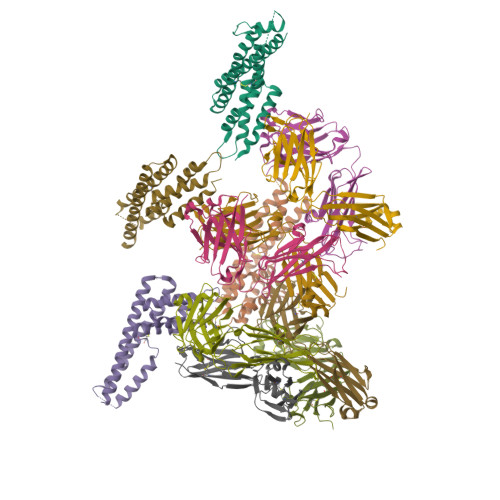
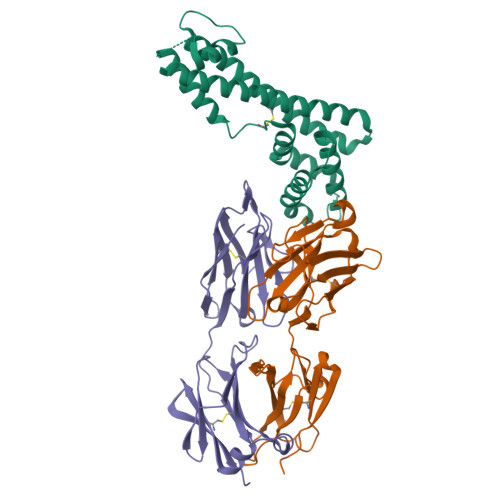
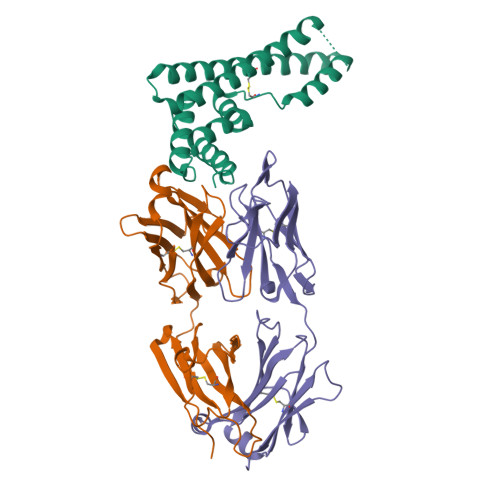
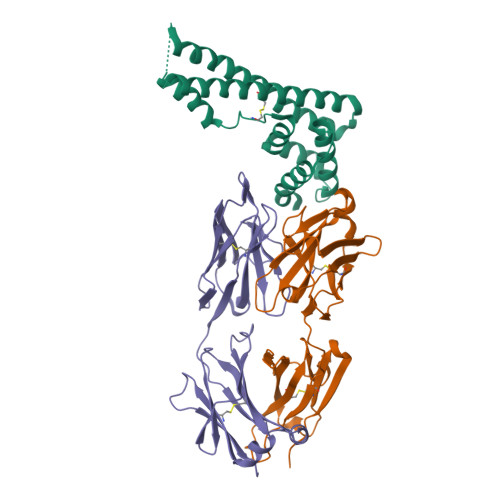
The structure of serum resistance-associated protein and its implications for human African trypanosomiasis.
Zoll, S., Lane-Serff, H., Mehmood, S., Schneider, J., Robinson, C.V., Carrington, M., Higgins, M.K.(2018) Nat Microbiol 3: 295-301
- PubMed: 29358741
- DOI: https://doi.org/10.1038/s41564-017-0085-3
- Primary Citation of Related Structures:
6ELU - PubMed Abstract:
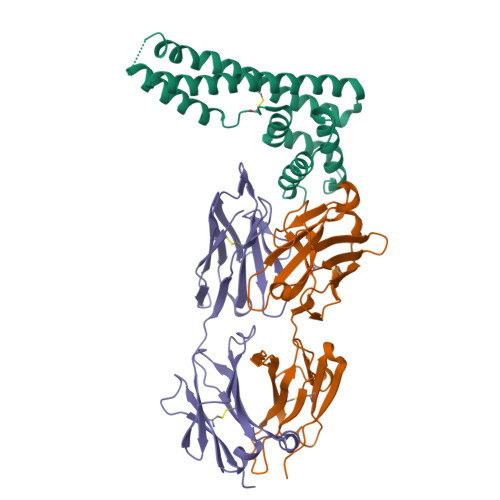
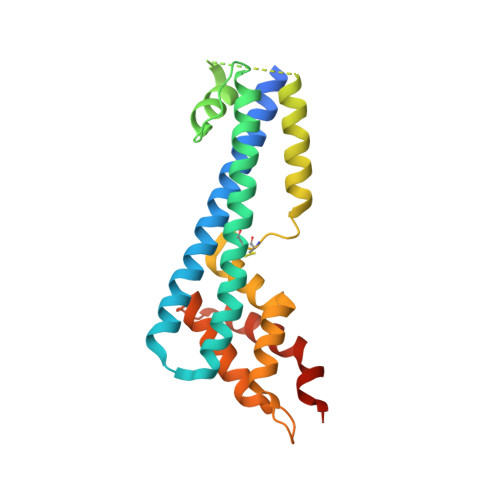
Only two trypanosome subspecies are able to cause human African trypanosomiasis. To establish an infection in human blood, they must overcome the innate immune system by resisting the toxic effects of trypanolytic factor 1 and trypanolytic factor 2 (refs. 1,2 ). These lipoprotein complexes contain an active, pore-forming component, apolipoprotein L1 (ApoL1), that causes trypanosome cell death 3 . One of the two human-infective subspecies, Trypanosoma brucei rhodesiense, differs from non-infective trypanosomes solely by the presence of the serum resistance-associated protein, which binds directly to ApoL1 and blocks its pore-forming capacity 3-5 . Since this interaction is the single critical event that renders T. b. rhodesiense human- infective, detailed structural information that allows identification of binding determinants is crucial to understand immune escape by the parasite. Here, we present the structure of serum resistance-associated protein and reveal the adaptations that occurred as it diverged from other trypanosome surface molecules to neutralize ApoL1. We also present our mapping of residues important for ApoL1 binding, giving molecular insight into this interaction at the heart of human sleeping sickness.
Organizational Affiliation:
Department of Biochemistry, University of Oxford, Oxford, UK.