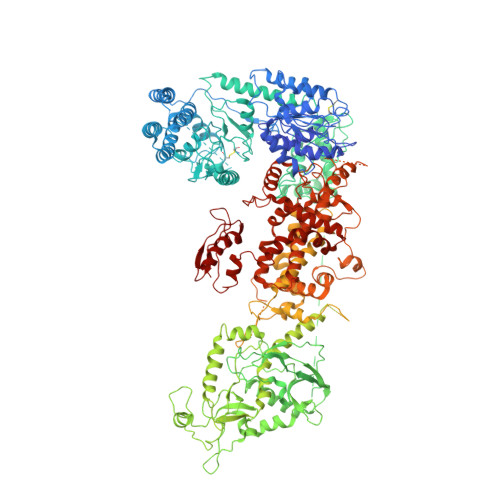
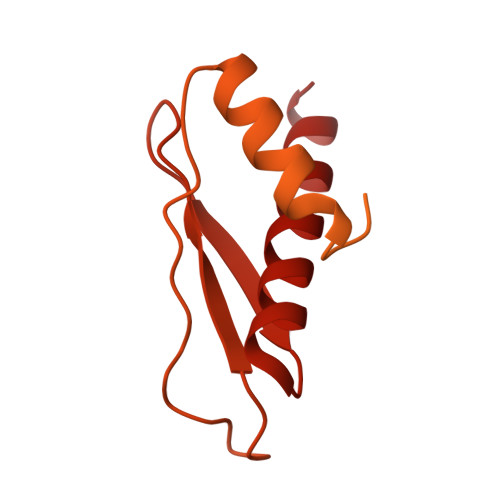
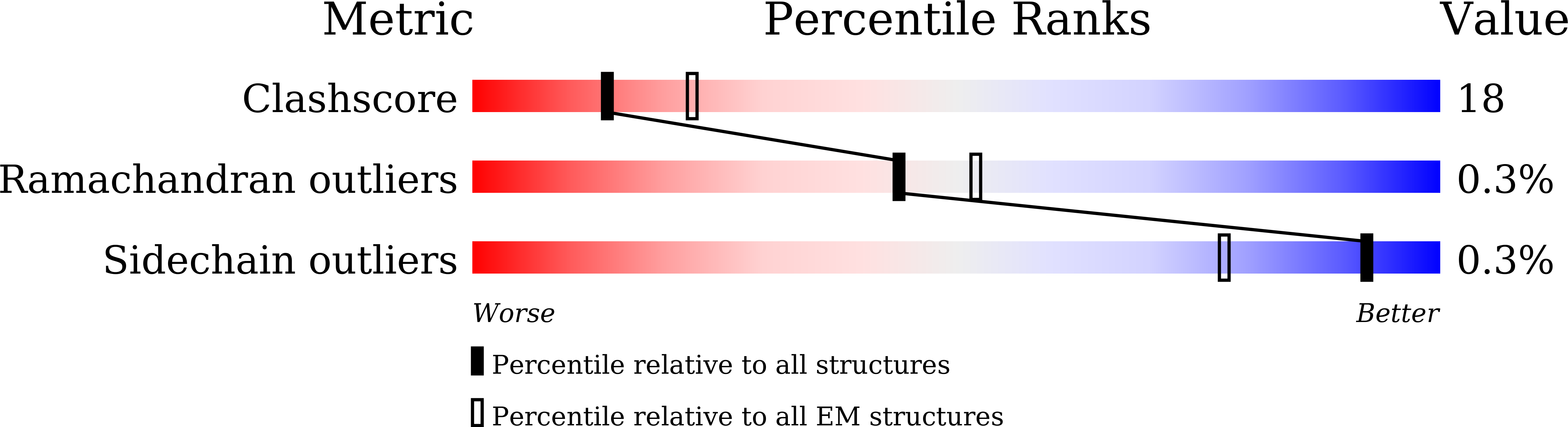Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate.
Liu, Z., Wang, J., Cheng, H., Ke, X., Sun, L., Zhang, Q.C., Wang, H.W.(2018) Cell 173: 1191-1203.e12
- PubMed: 29706542
- DOI: https://doi.org/10.1016/j.cell.2018.03.080
- Primary Citation of Related Structures:
5ZAK, 5ZAL, 5ZAM - PubMed Abstract:
Human Dicer (hDicer) is a multi-domain protein belonging to the RNase III family. It plays pivotal roles in small RNA biogenesis during the RNA interference (RNAi) pathway by processing a diverse range of double-stranded RNA (dsRNA) precursors to generate ∼22 nt microRNA (miRNA) or small interfering RNA (siRNA) products for sequence-directed gene silencing. In this work, we solved the cryoelectron microscopy (cryo-EM) structure of hDicer in complex with its cofactor protein TRBP and revealed the precise spatial arrangement of hDicer's multiple domains. We further solved structures of the hDicer-TRBP complex bound with pre-let-7 RNA in two distinct conformations. In combination with biochemical analysis, these structures reveal a property of the hDicer-TRBP complex to promote the stability of pre-miRNA's stem duplex in a pre-dicing state. These results provide insights into the mechanism of RNA processing by hDicer and illustrate the regulatory role of hDicer's N-terminal helicase domain.
Organizational Affiliation:
Ministry of Education Key Laboratory of Protein Sciences, Tsinghua-Peking Joint Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing, China 100084.