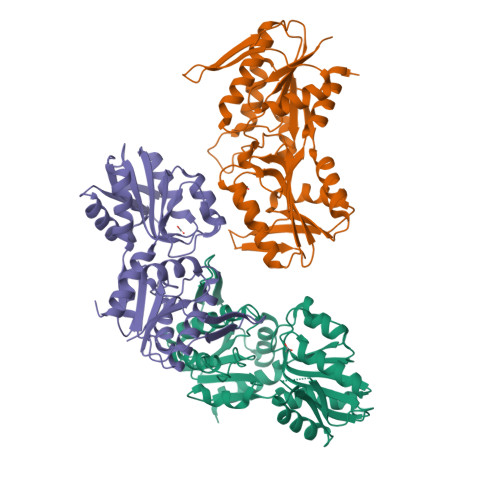
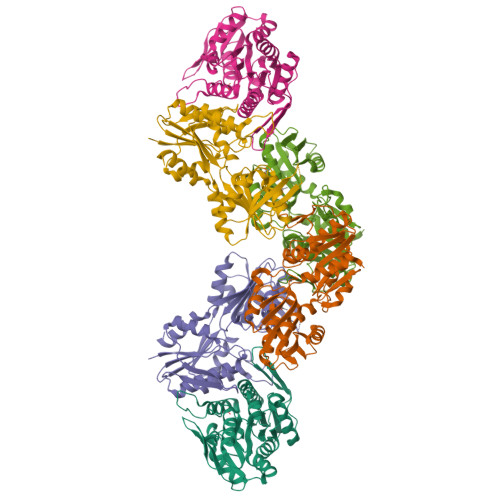
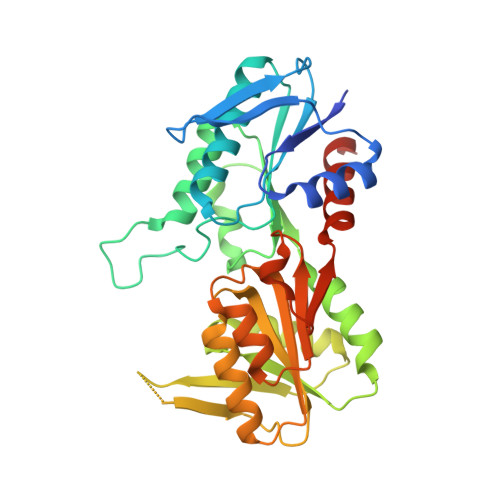
Biochemical and structural investigations on phosphoribosylpyrophosphate synthetase from Mycobacterium smegmatis.
Donini, S., Garavaglia, S., Ferraris, D.M., Miggiano, R., Mori, S., Shibayama, K., Rizzi, M.(2017) PLoS One 12: e0175815-e0175815
- PubMed: 28419153
- DOI: https://doi.org/10.1371/journal.pone.0175815
- Primary Citation of Related Structures:
5MP7 - PubMed Abstract:
Mycobacterium smegmatis represents one model for studying the biology of its pathogenic relative Mycobacterium tuberculosis. The structural characterization of a M. tuberculosis ortholog protein can serve as a valid tool for the development of molecules active against the M. tuberculosis target. In this context, we report the biochemical and structural characterization of M. smegmatis phosphoribosylpyrophosphate synthetase (PrsA), the ortholog of M. tuberculosis PrsA, the unique enzyme responsible for the synthesis of phosphoribosylpyrophosphate (PRPP). PRPP is a key metabolite involved in several biosynthetic pathways including those for histidine, tryptophan, nucleotides and decaprenylphosphoryl-arabinose, an essential precursor for the mycobacterial cell wall biosynthesis. Since M. tuberculosis PrsA has been validated as a drug target for the development of antitubercular agents, the data presented here will add to the knowledge of the mycobacterial enzyme and could contribute to the development of M. tuberculosis PrsA inhibitors of potential pharmacological interest.
Organizational Affiliation:
Department of Pharmaceutical Sciences, Università del Piemonte Orientale "A. Avogadro", Largo Donegani 2, Novara, Italy.