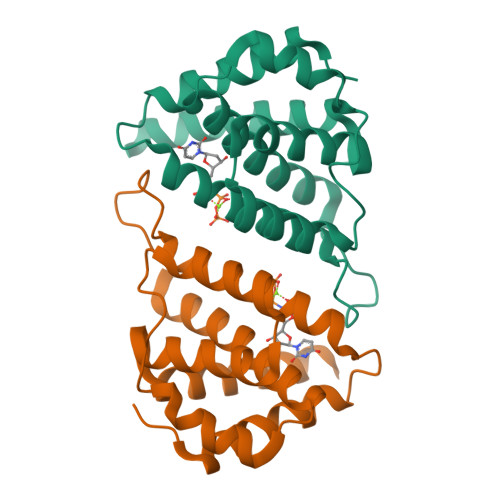
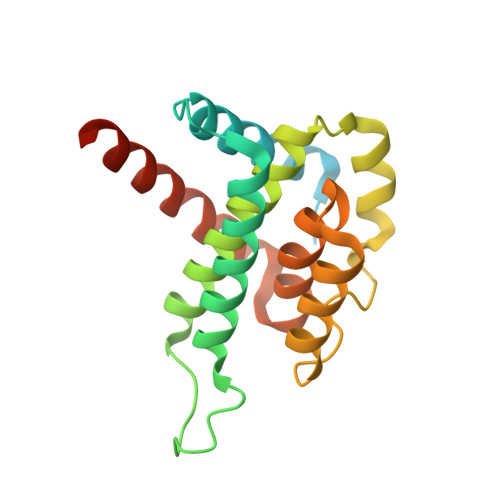
Pirating conserved phage mechanisms promotes promiscuous staphylococcal pathogenicity island transfer.
Bowring, J., Neamah, M.M., Donderis, J., Mir-Sanchis, I., Alite, C., Ciges-Tomas, J.R., Maiques, E., Medmedov, I., Marina, A., Penades, J.R.(2017) Elife 6
- PubMed: 28826473
- DOI: https://doi.org/10.7554/eLife.26487
- Primary Citation of Related Structures:
5MIL - PubMed Abstract:
Targeting conserved and essential processes is a successful strategy to combat enemies. Remarkably, the clinically important Staphylococcus aureus pathogenicity islands (SaPIs) use this tactic to spread in nature. SaPIs reside passively in the host chromosome, under the control of the SaPI-encoded master repressor, Stl. It has been assumed that SaPI de-repression is effected by specific phage proteins that bind to Stl, initiating the SaPI cycle. Different SaPIs encode different Stl repressors, so each targets a specific phage protein for its de-repression. Broadening this narrow vision, we report here that SaPIs ensure their promiscuous transfer by targeting conserved phage mechanisms. This is accomplished because the SaPI Stl repressors have acquired different domains to interact with unrelated proteins, encoded by different phages, but in all cases performing the same conserved function. This elegant strategy allows intra- and inter-generic SaPI transfer, highlighting these elements as one of nature's most fascinating subcellular parasites.
Organizational Affiliation:
Institute of Infection, Immunity and Inflammation, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow, United Kingdom.