Crystal Structure of Glyceraldehyde-3-Phosphate Dehydrogenase from the Gram-Positive Bacterial Pathogen A. vaginae, an Immunoevasive Factor that Interacts with the Human C5a Anaphylatoxin.
Querol-Garcia, J., Fernandez, F.J., Marin, A.V., Gomez, S., Fulla, D., Melchor-Tafur, C., Franco-Hidalgo, V., Alberti, S., Juanhuix, J., Rodriguez de Cordoba, S., Regueiro, J.R., Vega, M.C.(2017) Front Microbiol 8: 541-541
- PubMed: 28443070
- DOI: https://doi.org/10.3389/fmicb.2017.00541
- Primary Citation of Related Structures:
5LD5 - PubMed Abstract:
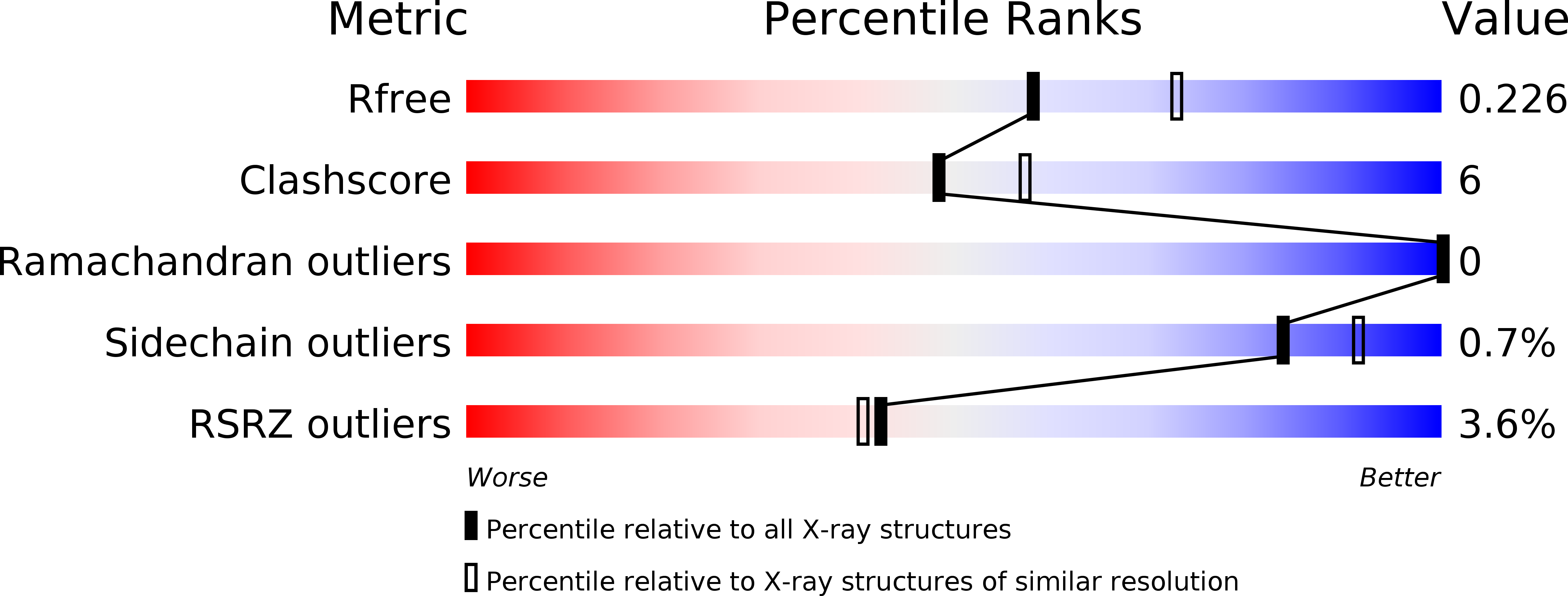
The Gram-positive anaerobic human pathogenic bacterium Atopobium vaginae causes most diagnosed cases of bacterial vaginosis as well as opportunistic infections in immunocompromised patients. In addition to its well-established role in carbohydrate metabolism, D-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from Streptococcus pyogenes and S. pneumoniae have been reported to act as extracellular virulence factors during streptococcal infections. Here, we report the crystal structure of GAPDH from A. vaginae ( Av GAPDH) at 2.19 Å resolution. The refined model has a crystallographic R free of 22.6%. Av GAPDH is a homotetramer wherein each subunit is bound to a nicotinamide adenine dinucleotide (NAD + ) molecule. The Av GAPDH enzyme fulfills essential glycolytic as well as moonlight (non-glycolytic) functions, both of which might be targets of chemotherapeutic intervention. We report that Av GAPDH interacts in vitro with the human C5a anaphylatoxin and inhibits C5a-specific granulocyte chemotaxis, thereby suggesting the participation of Av GAPDH in complement-targeted immunoevasion in a context of infection. The availability of high-quality structures of Av GAPDH and other homologous virulence factors from Gram-positive pathogens is critical for drug discovery programs. In this study, sequence and structural differences between Av GAPDH and related bacterial and eukaryotic GAPDH enzymes are reported in an effort to understand how to subvert the immunoevasive properties of GAPDH and evaluate the potential of Av GAPDH as a druggable target.
Organizational Affiliation:
Integrated Protein Science for Biomedicine & Biotechnology and Ciber de Enfermedades Raras, Center for Biological Research (CIB-CSIC)Madrid, Spain.