Germinal-center kinase-like kinase co-crystal structure reveals a swapped activation loop and C-terminal extension.
Marcotte, D., Rushe, M., M Arduini, R., Lukacs, C., Atkins, K., Sun, X., Little, K., Cullivan, M., Paramasivam, M., Patterson, T.A., Hesson, T., D McKee, T., May-Dracka, T.L., Xin, Z., Bertolotti-Ciarlet, A., Bhisetti, G.R., Lyssikatos, J.P., Silvian, L.F.(2017) Protein Sci 26: 152-162
- PubMed: 27727493
- DOI: https://doi.org/10.1002/pro.3062
- Primary Citation of Related Structures:
5J5T - PubMed Abstract:
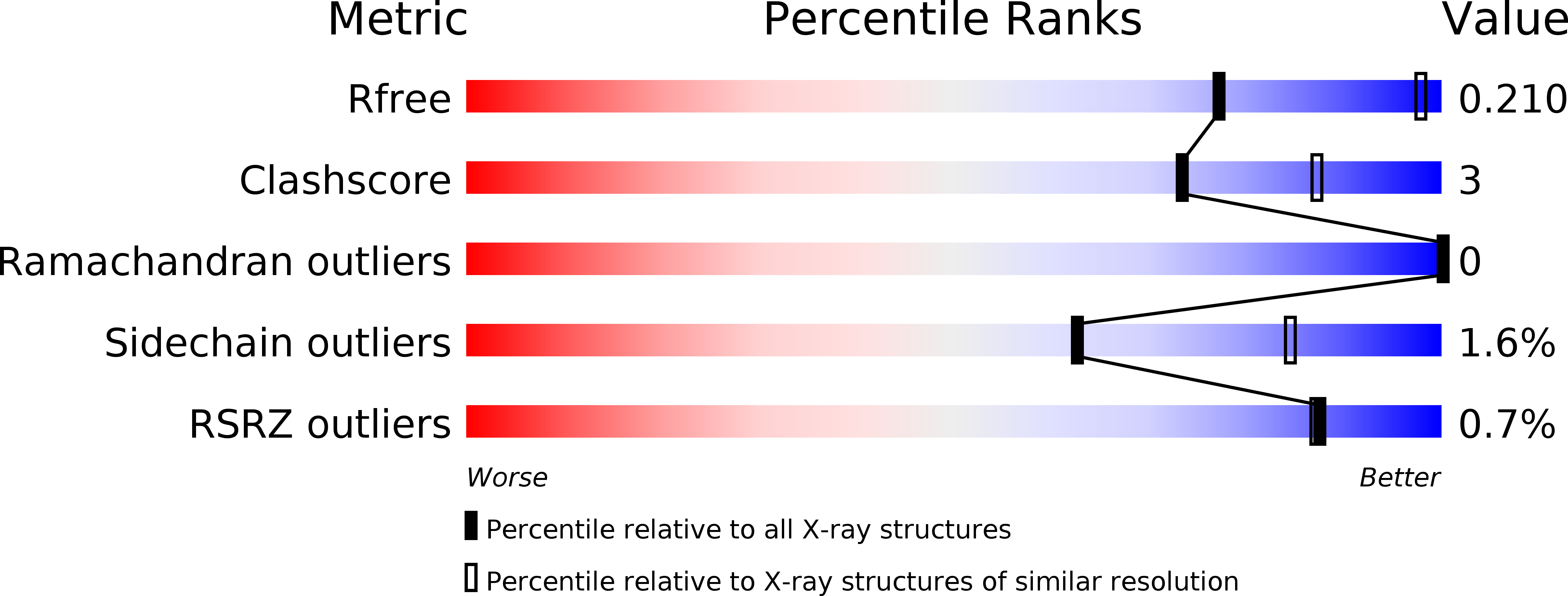
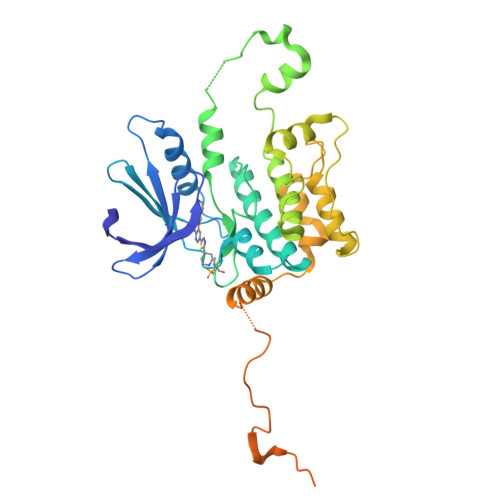
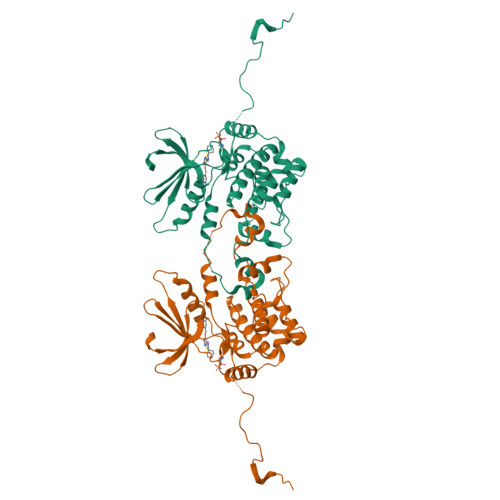
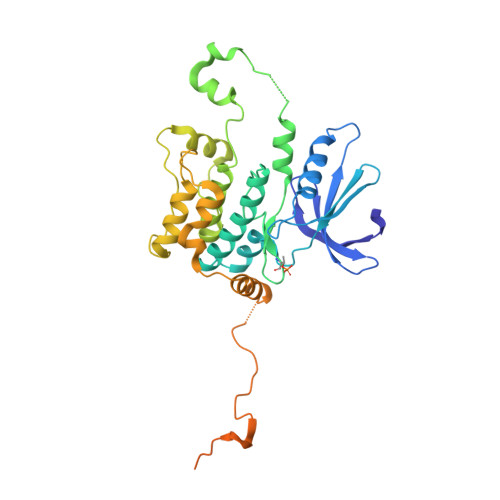
Germinal-center kinase-like kinase (GLK, Map4k3), a GCK-I family kinase, plays multiple roles in regulating apoptosis, amino acid sensing, and immune signaling. We describe here the crystal structure of an activation loop mutant of GLK kinase domain bound to an inhibitor. The structure reveals a weakly associated, activation-loop swapped dimer with more than 20 amino acids of ordered density at the carboxy-terminus. This C-terminal PEST region binds intermolecularly to the hydrophobic groove of the N-terminal domain of a neighboring molecule. Although the GLK activation loop mutant crystallized demonstrates reduced kinase activity, its structure demonstrates all the hallmarks of an "active" kinase, including the salt bridge between the C-helix glutamate and the catalytic lysine. Our compound displacement data suggests that the effect of the Ser170Ala mutation in reducing kinase activity is likely due to its effect in reducing substrate peptide binding affinity rather than reducing ATP binding or ATP turnover. This report details the first structure of GLK; comparison of its activation loop sequence and P-loop structure to that of Map4k4 suggests ideas for designing inhibitors that can distinguish between these family members to achieve selective pharmacological inhibitors.
Organizational Affiliation:
Department of Drug Discovery, Biogen Inc., 115 Binney Street, Cambridge, MA, 02142.