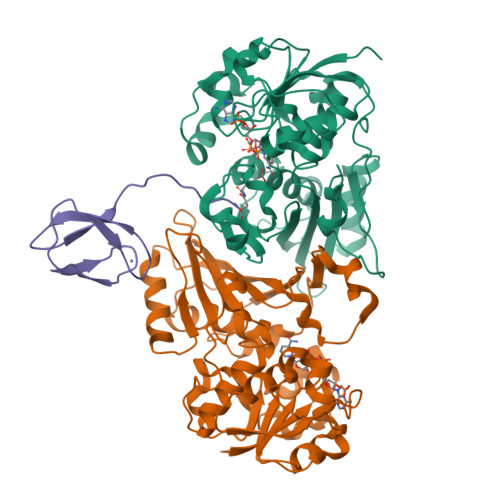
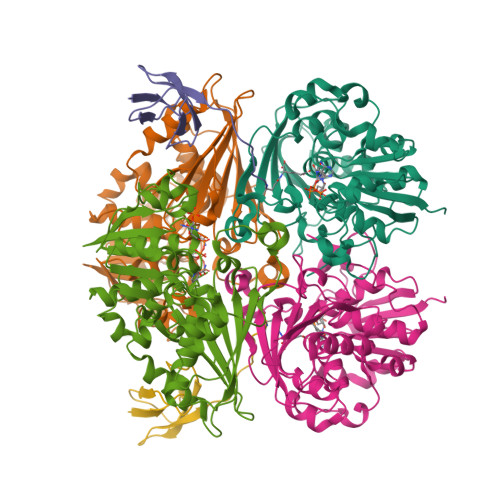
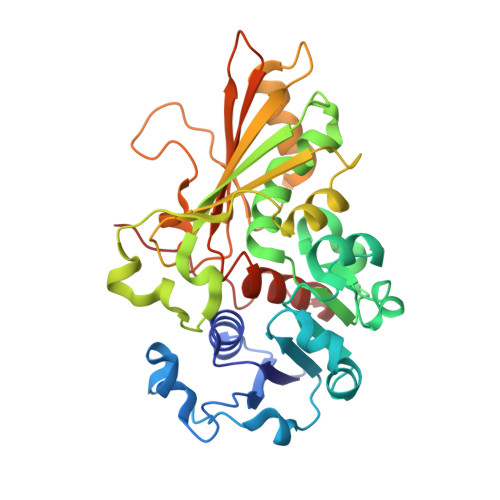
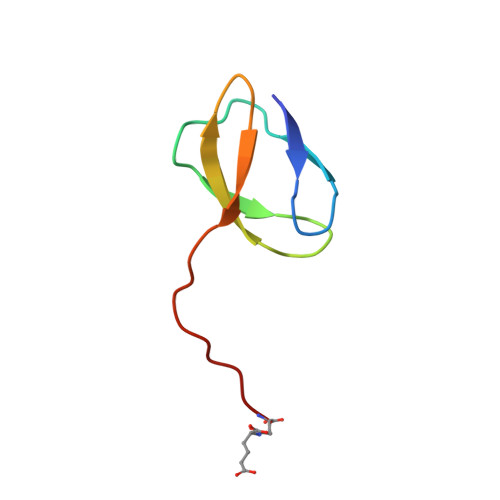
Crystal Structure of the LysYLysW Complex from Thermus thermophilus.
Shimizu, T., Tomita, T., Kuzuyama, T., Nishiyama, M.(2016) J Biological Chem 291: 9948-9959
- PubMed: 26966182
- DOI: https://doi.org/10.1074/jbc.M115.707034
- Primary Citation of Related Structures:
5EIN, 5EIO - PubMed Abstract:
Several bacteria and archaea utilize the amino group-carrier protein, LysW, for lysine biosynthesis, in which an isopeptide bond is formed between the C-terminal Glu of LysW and an amino group of α-aminoadipate (AAA). The resulting LysW-γ-AAA is phosphorylated by LysZ to form LysW-γ-AAA phosphate, which is subsequently reduced to LysW-γ-aminoadipic semialdehyde (LysW-γ-AASA) through a reaction catalyzed by LysY. In this study, we determined the crystal structures of LysY from Thermus thermophilus HB27 (TtLysY) complexed with TtLysW-γ-AASA and TtLysW-γ-AAA, respectively. In both structures, the globular domain of TtLysW was recognized by positively charged residues on helix α9 and the β11-α10 loop of TtLysY through conformational changes. A mutational analysis confirmed that the interactions observed between TtLysY and TtLysW are important for the function of TtLysY. The extended LysW recognition loop and conserved arginine residue were identified as signatures to discriminate LysY from ArgC, which is involved in arginine biosynthesis. Combined with the previously determined TtLysZ·TtLysW complex structure, TtLysW may simultaneously bind TtLysZ and TtLysY. These structural insights suggest the formation of a TtLysWZY ternary complex, in which the flexible C-terminal extension of TtLysW promotes the efficient transfer of the labile intermediate from the active site of TtLysZ to that of TtLysY during the sequential reactions catalyzed by TtLysZY.
Organizational Affiliation:
From the Biotechnology Research Center, University of Tokyo, 1-1-1, Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan.