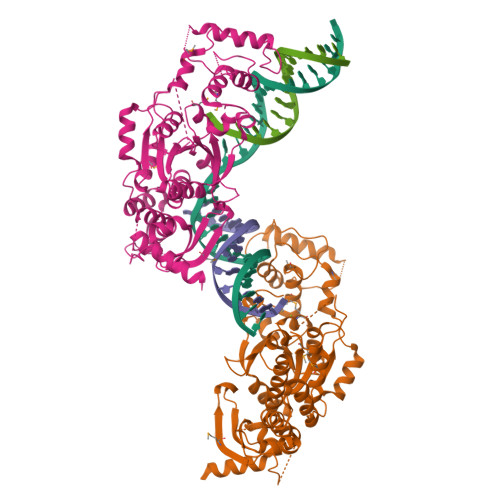
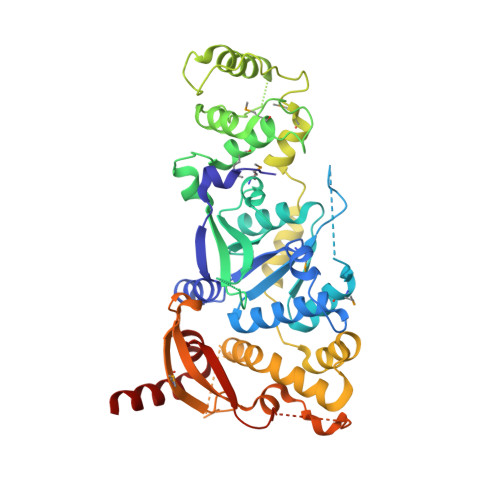
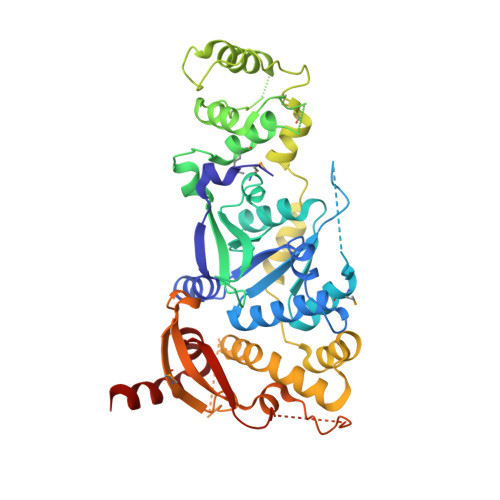
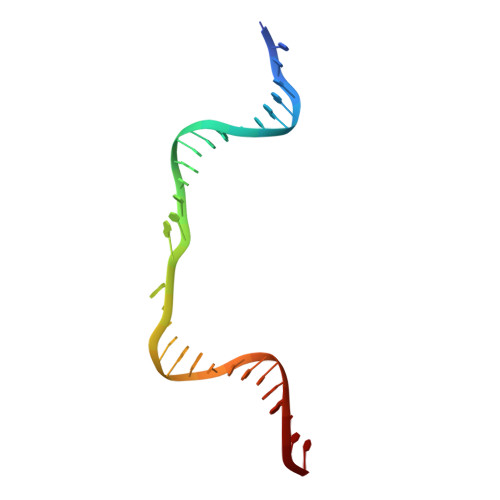
Crystal Structure of a Eukaryotic GEN1 Resolving Enzyme Bound to DNA.
Liu, Y., Freeman, A.D., Declais, A.C., Wilson, T.J., Gartner, A., Lilley, D.M.(2015) Cell Rep 13: 2565-2575
- PubMed: 26686639
- DOI: https://doi.org/10.1016/j.celrep.2015.11.042
- Primary Citation of Related Structures:
5CNQ, 5CO8 - PubMed Abstract:
We present the crystal structure of the junction-resolving enzyme GEN1 bound to DNA at 2.5 Å resolution. The structure of the GEN1 protein reveals it to have an elaborated FEN-XPG family fold that is modified for its role in four-way junction resolution. The functional unit in the crystal is a monomer of active GEN1 bound to the product of resolution cleavage, with an extensive DNA binding interface for both helical arms. Within the crystal lattice, a GEN1 dimer interface juxtaposes two products, whereby they can be reconnected into a four-way junction, the structure of which agrees with that determined in solution. The reconnection requires some opening of the DNA structure at the center, in agreement with permanganate probing and 2-aminopurine fluorescence. The structure shows that a relaxation of the DNA structure accompanies cleavage, suggesting how second-strand cleavage is accelerated to ensure productive resolution of the junction.
Organizational Affiliation:
Cancer Research UK Nucleic Acid Structure Research Group, MSI/WTB Complex, University of Dundee, Dow Street, Dundee DD1 5EH, UK.