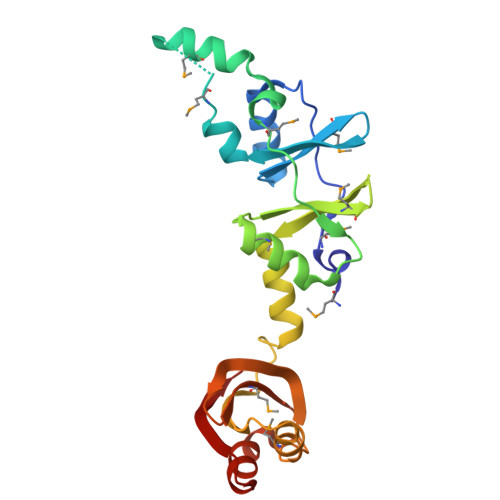
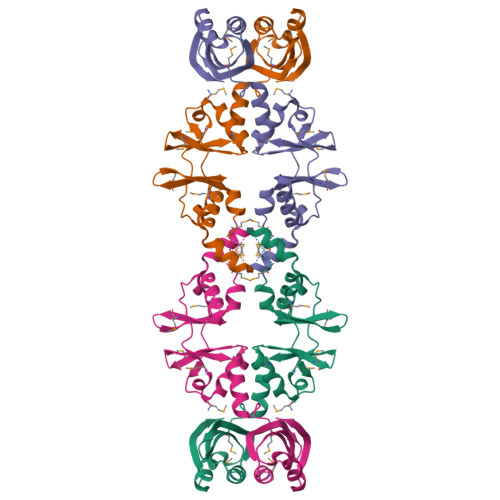
Crystal structure of a hypothetical protein, TTHA0829 from Thermus thermophilus HB8, composed of cystathionine-beta-synthase (CBS) and aspartate-kinase chorismate-mutase tyrA (ACT) domains.
Nakabayashi, M., Shibata, N., Ishido-Nakai, E., Kanagawa, M., Iio, Y., Komori, H., Ueda, Y., Nakagawa, N., Kuramitsu, S., Higuchi, Y.(2016) Extremophiles 20: 275-282
- PubMed: 26936147
- DOI: https://doi.org/10.1007/s00792-016-0817-y
- Primary Citation of Related Structures:
5AWE - PubMed Abstract:
TTHA0829 from Thermus thermophilus HB8 has a molecular mass of 22,754 Da and is composed of 210 amino acid residues. The expression of TTHA0829 is remarkably elevated in the latter half of logarithmic growth phase. TTHA0829 can form either a tetrameric or dimeric structure, and main-chain folding provides an N-terminal cystathionine-β-synthase (CBS) domain and a C-terminal aspartate-kinase chorismate-mutase tyrA (ACT) domain. Both CBS and ACT are regulatory domains to which a small ligand molecule can bind. The CBS domain is found in proteins from organisms belonging to all kingdoms and is observed frequently as two or four tandem copies. This domain is considered as a small intracellular module with a regulatory function and is typically found adjacent to the active (or functional) site of several enzymes and integral membrane proteins. The ACT domain comprises four β-strands and two α-helices in a βαββαβ motif typical of intracellular small molecule binding domains that help control metabolism, solute transport and signal transduction. We discuss the possible role of TTHA0829 based on its structure and expression pattern. The results imply that TTHA0829 acts as a cell-stress sensor or a metabolite acceptor.
Organizational Affiliation:
Graduate School of Life Science, University of Hyogo, 3-2-1 Koto, Kamigori-cho, Ako-gun, Hyogo, 678-1297, Japan. makoto-n.str@tmd.ac.jp.