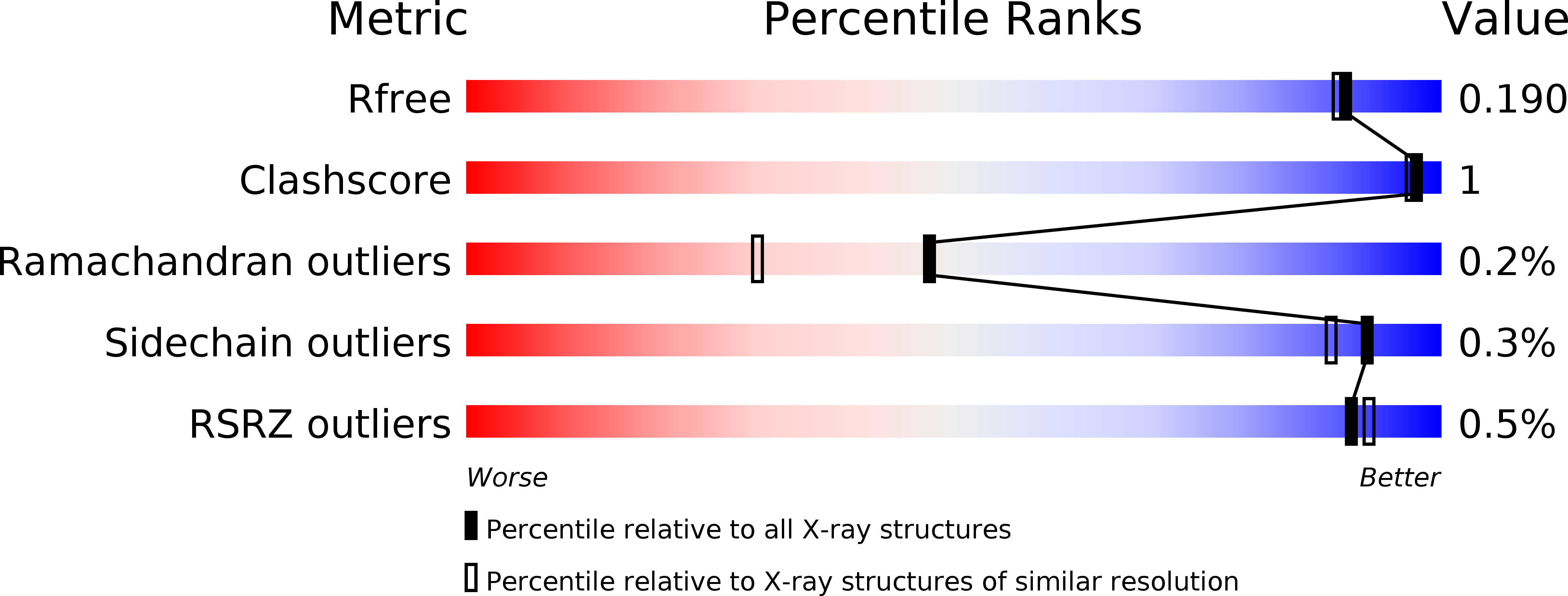
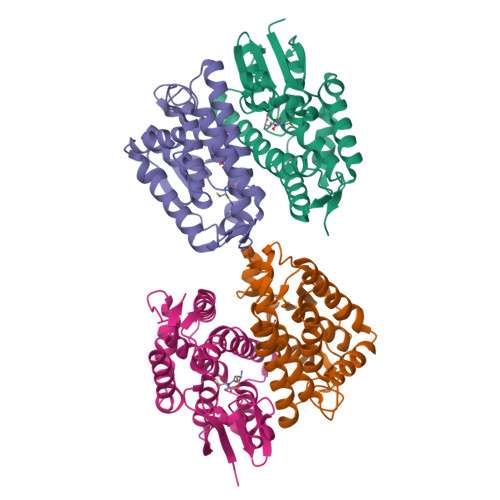
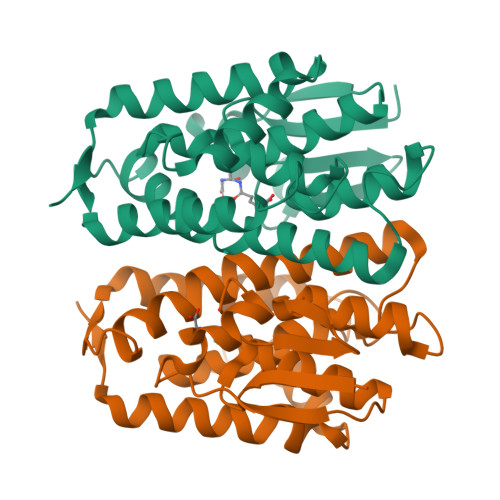
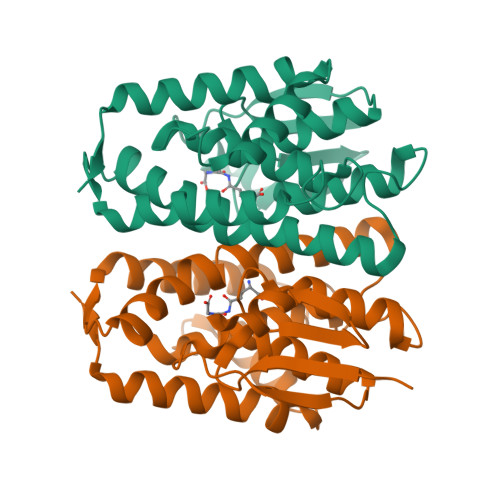
Epsilon glutathione transferases possess a unique class-conserved subunit interface motif that directly interacts with glutathione in the active site
Wongsantichon, J., Robinson, R.C., Ketterman, A.J.(2015) Biosci Rep 35
- PubMed: 26487708
- DOI: https://doi.org/10.1042/BSR20150183
- Primary Citation of Related Structures:
4YH2 - PubMed Abstract:
Epsilon class glutathione transferases (GSTs) have been shown to contribute significantly to insecticide resistance. We report a new Epsilon class protein crystal structure from Drosophila melanogaster for the glutathione transferase DmGSTE6. The structure reveals a novel Epsilon clasp motif that is conserved across hundreds of millions of years of evolution of the insect Diptera order. This histidine-serine motif lies in the subunit interface and appears to contribute to quaternary stability as well as directly connecting the two glutathiones in the active sites of this dimeric enzyme.
Organizational Affiliation:
Institute of Molecular and Cell Biology, A*STAR (Agency for Science, Technology and Research), Biopolis, Singapore 138673.